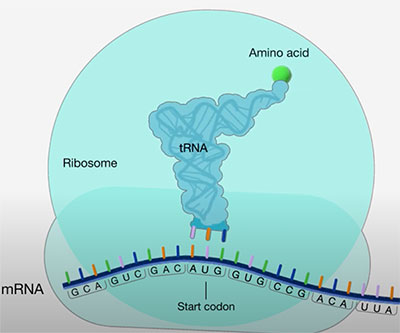
Illustration depicting the start of translation.
Credit: National Human Genome Research Institute
Translation—the reading of messenger RNA (mRNA) to make proteins—begins at a three-letter mRNA sequence called the start codon. Appropriate selection of the start codon by the cell’s translation machinery is crucial for proper protein synthesis. The efficiency of selection is influenced both by the mRNA sequence flanking the start codon and by levels of the translation initiation factors eIF1 and eIF5.
The Dever Lab and colleagues analyzed mammalian mRNA sequences to identify mRNAs with start codons in poor sequence contexts. They identified several Hox genes whose translation is sensitive to changes in the stringency of start codon selection. Hox genes encode proteins that specify the body plan of an embryo, ensuring that the correct structures form in the correct places of the body.
The researchers found five Hox genes in which the start codon for the main open reading frame (ORF)—the span of protein-defining genetic sequence between a start codon and a stop codon—is in evolutionarily conserved poor sequence context. Many mRNAs also contain upstream ORFs that encode short protein sequences that help regulate translation of the main ORF. The scientists identified an additional 13 Hox genes that contain evolutionarily conserved upstream ORFs with poor start codon sequence contexts.
The investigators conducted several experiments to evaluate the translation of these Hox genes when the stringency of start codon selection is altered—for example, by changing eIF1 and eIF5 levels. They found that translation of Hox genes with the main ORF start codon in a poor sequence context was reduced in conditions of high stringency. In contrast, translation of the Hox mRNAs with upstream ORF start codons in a poor sequence context was enhanced in conditions of high stringency. These findings illustrate how changes in start codon selection stringency have the potential to regulate the synthesis of specific proteins to control development.
Learn more about the Cell Regulation and Development Group: https://www.nichd.nih.gov/about/org/dir/affinity-groups/CRD
 BACK TO TOP
BACK TO TOP