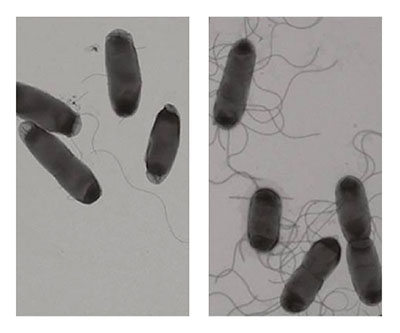
Escherichia coli cells without (left) and with (right) overexpression of the MotR small RNA, which increases flagella number.
Credit: Storz Lab, NICHD
A whip-like appendage called the flagellum helps bacteria move. The flagellum is composed of many proteins, the production of which is tightly regulated in response to environmental signals. Because producing flagellar components uses a great deal of energy, they are only made when motility is beneficial to the bacterium. Much is known about regulation of flagella synthesis at the level of transcription—the process of making an RNA copy of a gene’s DNA sequence—but relatively little is known about posttranscriptional regulation.
- Small RNAs (sRNAs) serve as key post-transcriptional regulators in bacteria. A recent study by the Storz Lab describes four sRNAs whose production is controlled by the flagella sigma factor σ28 in Escherichia coli. They found that the sRNAs target a wide range of genes, and three of the sRNAs affect flagella number and bacterial motility.
- Two of the sRNAs—MotR and FliX—have a unique mechanism of action and, respectively, act as an accelerator and a decelerator for flagella synthesis. MotR, expressed earlier in flagella growth, increases expression of flagellar and other proteins and flagella number. FliX, expressed later in growth, decreases expression of the proteins and flagella number.
- Overall, the study illustrates how posttranscriptional regulation by sRNAs helps enable nuanced control of flagella synthesis.
NICHD authors of the paper include Sahar Melamed, Aixia Zhang, Michal Jarnik, Joshua Mills, Hongen Zhang and Gisela Storz.
Learn more about the Cell and Structural Biology group: https://www.nichd.nih.gov/about/org/dir/affinity-groups/CSB
 BACK TO TOP
BACK TO TOP