Preprint: Single-cell analysis of shared signatures and transcriptional diversity during zebrafish development
March 21, 2023
SO proud to see this work out. This has been one of the major ongoing efforts in the lab for its first three years.
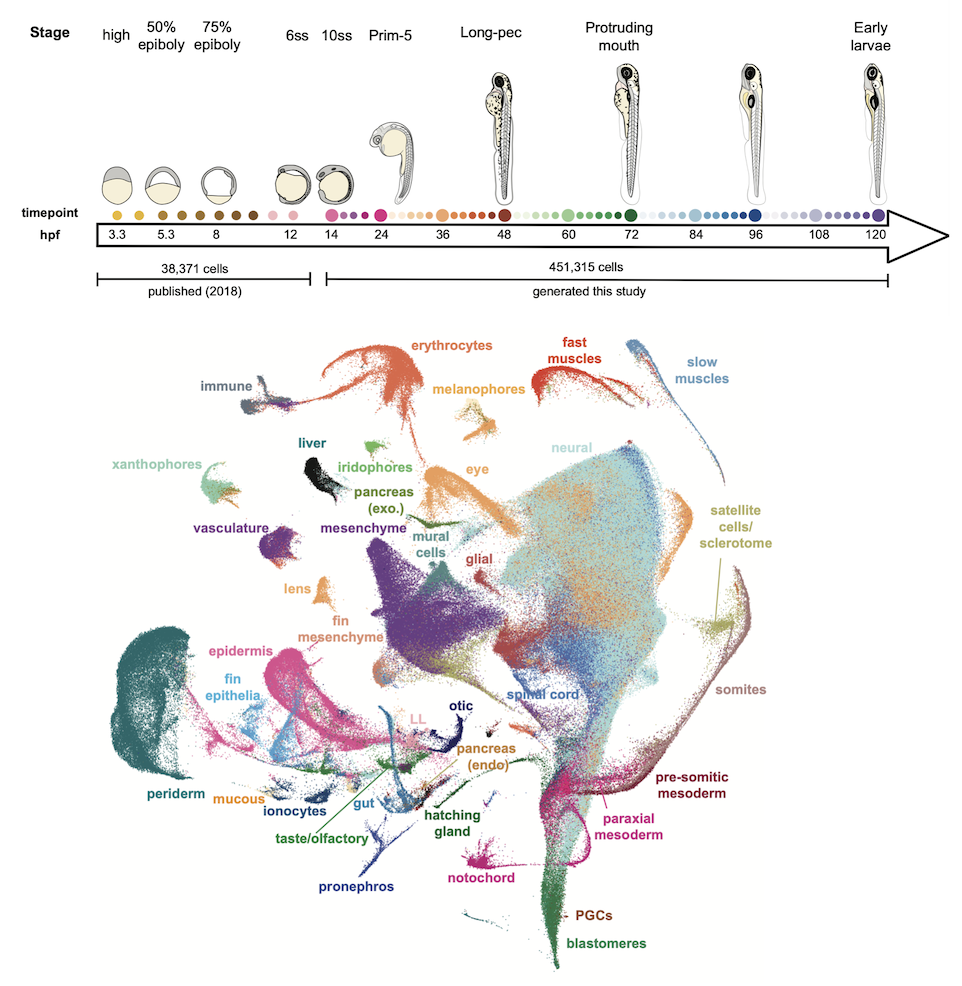
We generated a 489,000 cell atlas of 62 stages of zebrafish development that extends until the fish are freely swimming and hunting. Globally, we identified the long-term transcriptional states during development and the gene expression programs that are reused across cell types. Additionally, focused analyses within particular tissues revealed several surprising cell states, including distinct cell populations within intestinal smooth muscle, distinct pericyte subtypes, and zebrafish homologs of best4+ intestinal cells. We confirmed all populations using in situ hybridization, built transcriptional trajectories to describe their likely developmental process, and predicted candidate regulators of each cell type. To make this work accessible, we generated the online resource, Daniocell for browsing zebrafish single-cell RNAseq data.
Read it at bioRxiv!

 BACK TO TOP
BACK TO TOP