Within the past three decades, RNA-mediated regulation has been shown to be a significant mechanism for bacteria to quickly adapt to environmental stress conditions and finetune the expression of their genes. This post-transcriptional regulation is also critical for the infectivity of many pathogens, yet largely unstudied in the spirochete Borrelia burgdorferi, the causative agent of Lyme disease. Lyme disease is an emerging infectious disease and is the foremost vector-borne bacterial infection in the world.
The goal of the Adams lab is to better map the B. burgdorferi transcriptome, mechanistically characterize RNA regulators, and delineate RNAs critical for tick and mammalian infection.
Lyme disease is a multistage, debilitating bacterial infection, caused by the Borrelia burgdorferi sensu lato (Borreliella) group of spirochetes. The phylum Spirochaetes are most notably characterized by their long, slender, helical morphology and presence of periplasmic flagella, which are anchored on the cell-poles and span the length of the bacterium between inner and outer membranes. Swimming through their environment in corkscrew-like motions, spirochetes make up a small, unique, and pathogenic subset of bacteria.
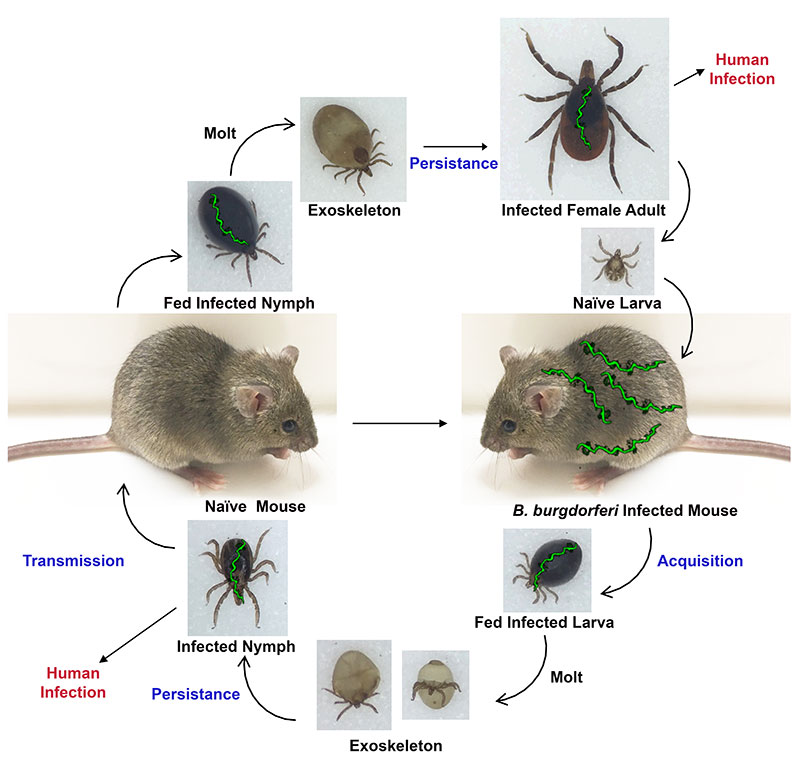
B. burgdorferi has a complex lifestyle in which the bacterium cycles between a tick vector (Ixodes scapularis) and various mammalian hosts on which it exclusively relies for transmission and nutrient acquisition. The reservoir of infected mammals (typically mice) and the transmission of B. burgdorferi by infected ticks leads to human infections - Lyme disease.
The genetics of B. burgdorferi is complex because the pathogen harbors a fragmented genome which is largely linear, unlike that of most prokaryotes. Additionally, the spirochete lacks the classical array of conserved bacterial metabolic genes and contains an unusually large percentage of unique genomic sequences specific to Borrelia species.
Transcriptional studies have demonstrated B. burgdorferi cells exhibit robust and dynamic changes in gene expression, varying with the bacteria’s environment and/or growth condition. However, the mechanisms responsible for mediating these observed differences have only been partially determined.
The Adams lab investigates gene regulation in B. burgdorferi, particularly post-transcriptional regulation. This includes classes of regulators such as (1) riboswitches which are encoded in the 5′ untranslated regions (UTRs) of mRNAs and bind specific ligands, often metabolic byproducts, to alter downstream gene expression (2) RNA thermometers, also 5′ encoded, which upon temperature changes alter their secondary structure to hide or expose ribosome binding sites (3) small upstream open reading frames (uORFs) whose translation affects the levels of the downstream mRNA and (4) base-pairing small RNAs (sRNAs) which function in trans to bind and regulate mRNA expression typically with RNA-binding proteins. We are also interested in better characterizing the bacteria’s transcriptome and regulatory events during tick and mammalian infection. Through this work we aim to better understand the biology of B. burgdorferi and the molecular mechanisms of its infectivity.
 BACK TO TOP
BACK TO TOP