DNA polymerases, 60 years of discoveries - Roger Woodgate Lab
1956
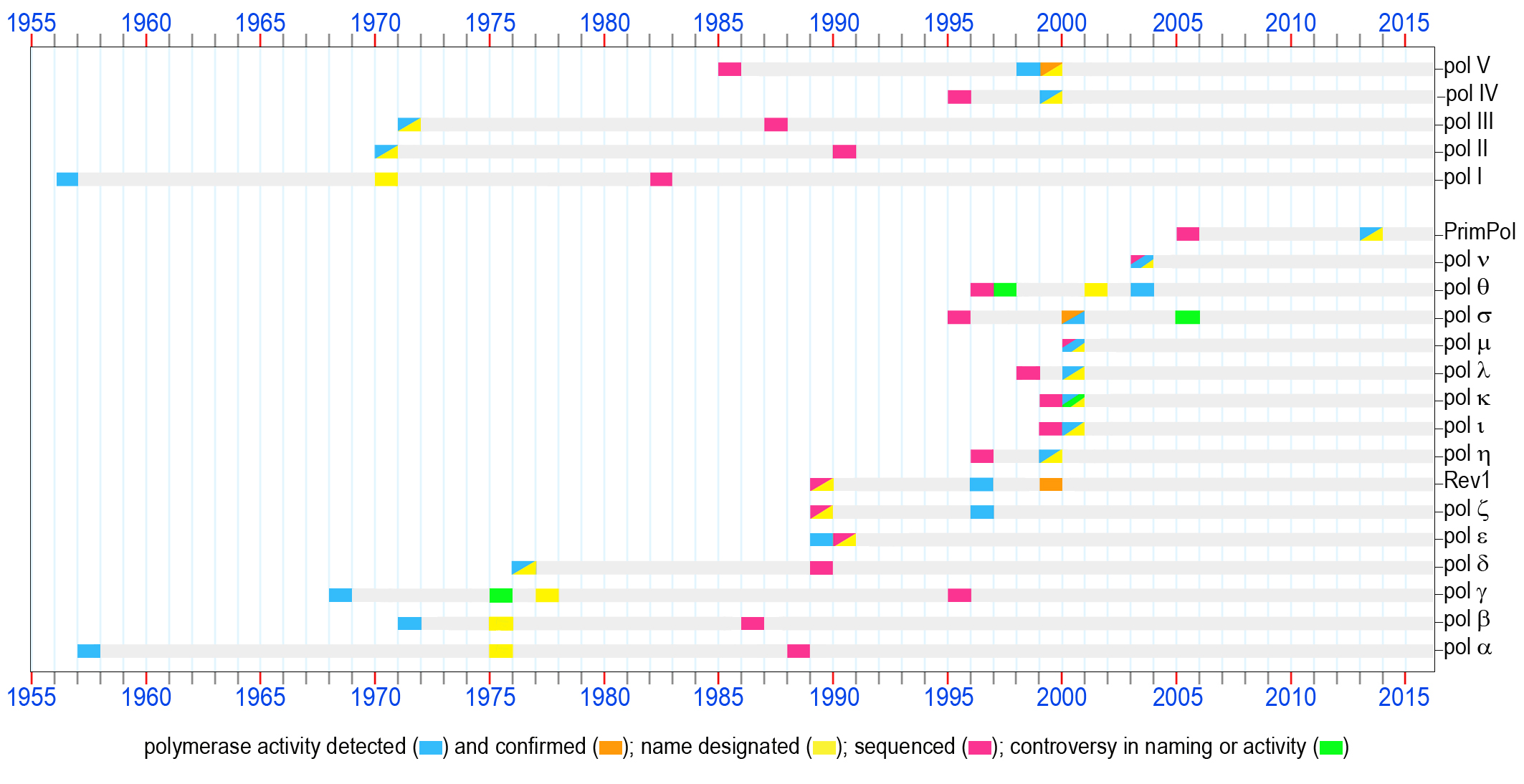
The first enzyme capable of copying DNA was discovered in E.coli extracts and was assumed at that time to be the only bacterial DNA polymerase [1]. Later, when a second E. coli DNA polymerase was purified, this enzyme playing an important role in prokaryotic DNA replication and repair was named pol I. The polA gene was sequenced in 1982 [2] (accession P00582a).
1957
The first eukaryotic DNA polymerase was identified [3]. When the uniform nomenclature was adopted in 1975, this enzyme was appropriately designated as pol α. Originally assumed to bear the sole responsibility for DNA synthesis in mammalian cells, this polymerase instead plays a key role in the initiation of chromosomal replication. The POLA gene was sequenced in 1988 [4] (accession CAA29920).
1968
An enzyme with DNA polymerase activity was isolated from the rat liver mitochondria [5, 6]. This enzyme, known since 1977 as pol g [7] is a major polymerase dealing with all transactions involving mitochondrial DNA in mammalian cells. The POLG gene was sequenced in 1995 [8] (accession CAA88012).
1970
A second DNA polymerase was discovered in E.coli by several groups in the USA and Germany [9-11]. According to the chronological order of discovery, it was named pol II. The sequencing of the polB gene was accomplished in 1990 [12] (accession P21189).
1971
During the course of purification of E.coli pol II, a third prokaryotic DNA polymerase was detected [13]. DNA polymerase III is now known to be the main prokaryotic replicative polymerase. The dnaE gene encoding the catalytic a-subunit of pol III was sequenced in 1987 [14] (accession P10443).
A second eukaryotic nuclear DNA polymerase later named pol β, was identified in mammalian cells and tissues practically simultaneously by several laboratories in the USA and England [15-19]. Very soon, it became apparent that this polymerase does not play a direct role in DNA replication. Instead, extensive research conducted by various groups revealed a major role for DNA pol β in base-excision repair. The POLB gene was sequenced in 1986 [20] (accession P06766).
1974
Aformal nomenclature designating each mammalian DNA polymerase with a Greek symbol was proposed in 1974 and accepted by attendees of the international conference on eukaryotic DNA polymerases in 1975 [21].According to the establishednomenclature, the first two mammalian DNA polymerases were designated pols α and β. It is worth noting that the third DNA polymerase which was given the name pol γ was and a forth DNA polymerase found in mitochondria (designated DNA polymerase-mt) were later shown to be identical and the name pol γ has been retained for this mitochondrial polymerase [7].
1976
The first nuclear polymerase containing an associated 3'®5' exonuclease activity was purified and called pol δ [22]. Later, this polymerase was shown to be an essential component of the eukaryotic replication machinery. The sequencing of the POLD1 gene was accomplished in 1989 [23] (accession P15436).
1987
It was proposed that DNA polymerases should be classified into discrete families based on their evolutionary relatedness. The first two evolutionary groups of DNA polymerases were designated as polymerase families A- and B- according to the amino acid homology to E coli pols I and II, respectively [24]. In 1991 two additional groups typified by the catalytic subunit of E coli pols III and eukaryotic pol β were designated as families C- and X-, respectively [25]. In 1999 family D- was proposed to group polymerases involved in the DNA replication machinery of the Euryarchaeota [26]. In 2001, proteins originally defined as belonging to the UmuC/DinB/Rev1/Rad30 superfamily and involved in mutagenesis and TLS DNA synthesis were designated as Y-family polymerases [27].
1989
The fourth nuclear DNA polymerase in mammalian cells, pol ε, was first reported as a PCNA-independent form of pol δ [28]. Subsequently, this enzyme was recognized as a distinct DNA polymerase and accordingly it was named pol ε. Similar to pol δ, pol ε is equipped with 3'®5' exonuclease proofreading activity and is essential for replication of the eukaryotic genome. The POLE1 gene was sequenced in 1990 [29] (accession P21951).
1996
Saccharomyces cerevisiae DNA pol ζ was characterized as a complex of Rev3 and Rev7 proteins [30]. These studies confirmed the hypothesis that the Rev3 gene long known to be involved in damage-induced and spontaneous mutagenesis, encodes the first DNA polymerase specializing in TLS [31] (accession P14284). This prediction was made based on the homology of Rev3 to other genes encoding B-family DNA polymerases.
In the same year, the same group discovered dCMP transferase activity for the S. cerevisiae REV1 protein [32] that was know at the time to be required for the damage-induced mutagenesis and having ~25% identity with the E.coli UmuC protein [33] (accession P12689). It was proposed that the CMP transferase function was important for mutagenic TLS involving pol ζ. However, Rev1 was not recognized as belonging to the broad superfamily of DNA-dependent DNA polymerases until 1999, when deoxynucleotidyl transferase activity was detected in several enzymes homologous to REV1.
1998
The first evidence suggesting that the E. coli UmuD'2C complex consisting of the umuDC gene products [34] [35] (accession P04152), s a DNA polymerase was demonstrated [36]. At that time, it was shown that in vitro the UmuD'2C complex could copy an abasic site-containing DNA template without the assistance of any other polymerase, although the possibility of contamination with trace amounts of other DNA polymerase were not entirely ruled out. A year later, additional biochemical studies unequivocally confirmed that the UmuD'2C complex is a bona fide DNA polymerase designated as E.coli pol V [37].
1999
The S.cerevisiae RAD30 gene which was previously identified by sequence homology to prokaryotic UmuC and DinB in 1996 [38, 39] was shown to encode DNA polymerase η [40]. Shortly thereafter, human polymerase η was characterized [41]. Polη became one of the founding members of the new Y-family of DNA polymerases [27]. Nonsense, or frameshift mutations in the gene (RAD30A, POLH, XPV) encoding pol ηare responsible for the Xeroderm Pigmentosum Variant syndrome in humans [41, 42].
The same week that the DNA polymerase activity of the UmuD'2C encoded pol V was confirmed, a manuscript describing the TLS activity of E.coli DinB was published [43]. This polymerase became known as E.coli DNA pol IV. The dinB gene was originally identified as dinP in 1995 [44] (accession BAA07593) and despite being shown to be allelic with dinB in 1999 the name of dinP, rather than the correct name of dinB, is still often used in Genbank data files describing related proteins.
2000
The TLS activity of two eukaryotic Y-family polymerases ι [45] and κ [46] (products of POLI (RAD30B) and POLK (DINB1) genes, respectively) was demonstrated a year after they were cloned [47, 48] [accession numbers AAD50381 (pol ι) and AAF02541 pol κ)].
Three X-family polymerases implicated in participating in different types of DNA transactions (such as BER, non-homologous end joining repair, V(D)J recombination, TLS, and sister chromatid cohesion) were discovered. This includes pol λ [49] (the sequence was first submitted in 1998 [50] [accession number CAB65241]), pol μ [51] (accession CAB65075), and pol σ [52] (first sequenced in 1995 [53] accession P53632). It should be noted that the DNA polymerase activity of pol σ has been contested by Haracska et al., [54], who suggest that the protein is actually a poly-A RNA polymerase, rather than a bona fide DNA polymerase.
2003
Two mammalian A-family DNA polymerases that are homologous to the DNA cross-link sensitivity protein Mus308 and implicated in different defense pathways against DNA damage were identified and characterized; pol q [55] and pol ν [56]. It worth mentioning that pol q, the only polymerase known to contain a helicase domain, was first identified in 1997 in the genomes of a variety of eukaryotic organisms based on sequence homology to E. coli DNA pol I [57-59] (accession numbers AAB67306 and AAC33565). The polymerase was originally named pol η [60], but was later renamed pol q [61]. Pol ν was sequenced in 2003 [56] (accession NP_861524).
2013
An ability to replicate both damaged and undamaged DNA templates was detected in PrimPol, an enzyme belonging to the archaeal-eukaryotic primase superfamily [62, 63]. The gene encoding PrimPol was first sequenced in 2005 [64] (accession NP_689896). The ability to catalyze TLS is only one of the broad enzymatic activities of the PrimPol enzymes that have been implicated in a large variety of cellular functions.
a GeneBank Sequence identifiers are listed [65].
References
- Kornberg, A., et al., Enzymic synthesis of deoxyribonucleic acid. Biochim Biophys Acta, 1956. 21(1): p. 197-8.
- Joyce, C.M., W.S. Kelley, and N.D. Grindley, Nucleotide sequence of the Escherichia coli polA gene and primary structure of DNA polymerase I. J Biol Chem, 1982. 257(4): p. 1958-64.
- Bollum, F.J. and V.R. Potter, Thymidine incorporation of into deoxyribonucleic acid of rat liver homogenates. J. Am. Chem. Soc., 1957. 79(13): p. 3603–3604.
- Wong, S.W., et al., Human DNA polymerase α gene expression is cell proliferation dependent and its primary structure is similar to both prokaryotic and eukaryotic replicative DNA polymerases. EMBO J, 1988. 7(1): p. 37-47.
- Kalf, G.F. and J.J. Ch'ih, Purification and properties of deoxyribonucleic acid polymerase from rat liver mitochondria. J Biol Chem, 1968. 243(18): p. 4904-16.
- Meyer, R.R. and M.V. Simpson, DNA biosynthesis in mitochondria: partial purification of a distinct DNA polymerase from isolated rat liver mitochondria. Proc Natl Acad Sci U S A, 1968. 61(1): p. 130-7.
- Bolden, A., G.P. Noy, and A. Weissbach, DNA polymerase of mitochondria is a γ-polymerase. J Biol Chem, 1977. 252(10): p. 3351-6.
- Ropp, P.A. and W.C. Copeland, Characterization of a new DNA polymerase from Schizosaccharomyces pombe: a probable homologue of the Saccharomyces cerevisiae DNA polymerase λ. Gene, 1995. 165(1): p. 103-7.
- Knippers, R., DNA polymerase II. Nature, 1970. 228: p. 1050-1053.
- Kornberg, T. and M.L. Gefter, DNA synthesis in cell-free extracts of a DNA polymerase-defective mutant. Biochem Biophys Res Commun, 1970. 40(6): p. 1348-55.
- Moses, R.E. and C.C. Richardson, A new DNA polymerase activity of Escherichia coli. I. Purification and properties of the activity present in E. coli polA1. Biochem Biophys Res Commun, 1970. 41(6): p. 1557-64.
- Chen, H., et al., Nucleotide sequence and deletion analysis of the polB gene of Escherichia coli. DNA Cell Biol, 1990. 9(9): p. 631-5.
- Kornberg, T. and M.L. Gefter, Purification and DNA synthesis in cell-free extracts: properties of DNA polymerase II. Proc Natl Acad Sci USA, 1971. 68: p. 761-764.
- Tomasiewicz, H.G. and C.S. McHenry, Sequence analysis of the Escherichia coli dnaE gene. J Bacteriol, 1987. 169: p. 5735-5744.
- Weissbach, A., et al., DNA polymerases from human cells. Nat New Biol, 1971. 231(23): p. 167-70.
- Chang, L.M. and F.J. Bollum, Low molecular weight deoxyribonucleic acid polymerase in mammalian cells. J Biol Chem, 1971. 246(18): p. 5835-7.
- Baril, E.F., et al., Deoxyribonucleic acid polymerase with rat liver ribosomes and smooth membranes. Purification and properties of the enzymes. Biochemistry, 1971. 10(11): p. 1981-92.
- Haines, M.E., A.M. Holmes, and I.R. Johnston, Distinct cytoplasmic and nuclear DNA polymerases from rat liver. FEBS Lett, 1971. 17(1): p. 63-67.
- Berger, H., Jr., R.C. Huang, and J.L. Irvin, Purification and characterization of a deoxyribonucleic acid polymerase from rat liver. J Biol Chem, 1971. 246(23): p. 7275-83.
- Zmudzka, B.Z., et al., Structure of rat DNA polymerase β revealed by partial amino acid sequencing and cDNA cloning. Proc Natl Acad Sci U S A, 1986. 83(14): p. 5106-10.
- Weissbach, A., et al., Nomenclature of eukaryotic DNA polymerases. Science, 1975. 190(4212): p. 401-2.
- Byrnes, J.J., et al., A new mammalian DNA polymerase with 3' to 5' exonuclease activity: DNA polymerase δ. Biochemistry, 1976. 15(13): p. 2817-23.
- Boulet, A., et al., Structure and function of the Saccharomyces cerevisiae CDC2 gene encoding the large subunit of DNA polymerase III. EMBO J, 1989. 8(6): p. 1849-54.
- Jung, G.H., et al., Bacteriophage PRD1 DNA polymerase: evolution of DNA polymerases. Proc Natl Acad Sci U S A, 1987. 84(23): p. 8287-91.
- Ito, J. and D.K. Braithwaite, Compilation and alignment of DNA polymerase sequences. Nucleic Acids Res., 1991. 19: p. 4045-4057.
- Cann, I.K. and Y. Ishino, Archaeal DNA replication: identifying the pieces to solve a puzzle. Genetics, 1999. 152: p. 1249-1267.
- Ohmori, H., et al., The Y-family of DNA polymerases. Mol Cell, 2001. 8: p. 7-8.
- Focher, F., et al., Calf thymus DNA polymerase δ independent of proliferating cell nuclear antigen (PCNA). Nucleic Acids Res, 1989. 17(5): p. 1805-21.
- Morrison, A., et al., A third essential DNA polymerase in S. cerevisiae. Cell, 1990. 62(6): p. 1143-51.
- Nelson, J.R., C.W. Lawrence, and D.C. Hinkle, Thymine-thymine dimer bypass by yeast DNA polymerase ζ. Science, 1996. 272: p. 1646-1649.
- Morrison, A., et al., REV3, a Saccharomyces cerevisiae gene whose function is required for induced mutagenesis, is predicted to encode a nonessential DNA polymerase. J Bacteriol, 1989. 171(10): p. 5659-67.
- Nelson, J.R., C.W. Lawrence, and D.C. Hinkle, Deoxycytidyl transferase activity of yeast REV1 protein. Nature, 1996. 382: p. 729-731.
- Larimer, F.W., J.R. Perry, and A.A. Hardigree, The REV1 gene of Saccharomyces cerevisiae: isolation, sequence and functional analysis. J Bacteriol, 1989. 171(1): p. 230-237.
- Perry, K.L., et al., umuDC and mucAB operons whose products are required for UV light and chemical-induced mutagenesis: UmuD, MucA, and LexA products share homology. Proc Natl Acad Sci U S A, 1985. 82: p. 4331-4335.
- Kitagawa, Y., et al., Structural analysis of the umu operon required for inducible mutagenesis in Escherichia coli. Proc Natl Acad Sci USA, 1985. 82: p. 4336-4340.
- Tang, M., et al., Biochemical basis of SOS-induced mutagenesis in Escherichia coli: reconstitution of in vitro lesion bypass dependent on the UmuD'2C mutagenic complex and RecA. Proc Natl Acad Sci U S A, 1998. 95: p. 9755-9760.
- Tang, M., et al., UmuD'2C is an error-prone DNA polymerase, Escherichia coli, DNA pol V. Proc Natl Acad Sci U S A, 1999. 96: p. 8919-8924.
- Kulaeva, O.I., et al., Identification of a DinB/UmuC homolog in the archeon Sulfolobus solfataricus. Mutation Research, 1996. 357: p. 245-253.
- McDonald, J.P., A.S. Levine, and R. Woodgate, The Saccharomyces cerevisiae RAD30 gene, a homologue of Escherichia coli dinB and umuC, is DNA damage inducible and functions in a novel error-free postreplication repair mechanism. Genetics, 1997. 147: p. 1557-1568.
- Johnson, R.E., S. Prakash, and L. Prakash, Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase, polη. Science, 1999. 283: p. 1001-1004.
- Masutani, C., et al., The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase η. Nature, 1999. 399: p. 700-704.
- Johnson, R.E., et al., hRAD30 mutations in the variant form of Xeroderma Pigmentosum. Science, 1999. 285: p. 263-265.
- Wagner, J., et al., The dinB gene encodes an novel Escherichia coli DNA polymerase (DNA pol IV) involved in mutagenesis. Mol Cell, 1999. 4: p. 281-286.
- Ohmori, H., et al., dinP, a new gene in Escherichia coli, whose product shows similarities to UmuC and its homologues. Mutation Research, 1995. 347: p. 1-7.
- Tissier, A., et al., polι, a remarkably error-prone human DNA polymerase. Genes & Dev, 2000. 14: p. 1642-1650.
- Ohashi, E., et al., Fidelity and processivity of DNA synthesis by DNA polymerase k, the product of the human DINB1 gene. J Biol Chem, 2000. 275: p. 39786-39684.
- McDonald, J.P., et al., Novel human and mouse homologs of Saccharomyces cerevisiae DNA polymerase η. Genomics, 1999. 60: p. 20-30.
- Gerlach, V.L., et al., Human and mouse homologs of Escherichia coli DinB (DNA polymerase IV), members of the UmuC/DinB superfamily. Proc Natl Acad Sci U S A, 1999. 96: p. 11922-11927.
- Garcia-Diaz, M., et al., DNA polymerase lambda (Pol l), a novel eukaryotic DNA polymerase with a potential role in meiosis. J Mol Biol, 2000. 301: p. 851-867.
- Blanco, L., DNA polymerase lambda [Mus musculus]. 1998.
- Dominguez, O., et al., DNA polymerase mu (Pol m), homologous to TdT, could act as a DNA mutator in eukaryotic cells. EMBO J., 2000. 19: p. 1731-1742.
- Wang, Z., et al., Pol κ: a DNA polymerase required for sister chromatid cohesion. Science, 2000. 289: p. 774-779.
- Sadoff, B.U., et al., Isolation of mutants of Saccharomyces cerevisiae requiring DNA topoisomerase I. Genetics, 1995. 141(2): p. 465-79.
- Haracska, L., et al., Trf4 and Trf5 proteins of Saccharomyces cerevisiae exhibit poly(A) RNA polymerase activity but no DNA polymerase activity. Mol Cell Biol, 2005. 25(22): p. 10183-9.
- Seki, M., F. Marini, and R.D. Wood, Polθ (Pol theta), a DNA polymerase and DNA-dependent ATPase in human cells. Nucleic Acids Res, 2003. 31(21): p. 6117-26.
- Marini, F., et al., Polν, a nuclear PolA family DNA polymerase homologous to the DNA cross-link sensitivity protein Mus308. J Biol Chem, 2003. 278(34): p. 32014-9.
- Harris, P.V., et al., Molecular cloning of Drosophila mus308, a gene involved in DNA cross-link repair with homology to prokaryotic DNA polymerase I genes. Mol Cell Biol, 1996. 16(10): p. 5764-71.
- Sonnhammer, E.L. and J.C. Wootton, Widespread eukaryotic sequences, highly similar to bacterial DNA polymerase I, looking for functions. Curr Biol, 1997. 7(8): p. R463-5.
- Sharief, F.S., et al., Cloning and chromosomal mapping of the human DNA polymerase q (POLQ), the eighth human DNA polymerase. Genomics, 1999. 59: p. 90-96.
- Burtis, K.C. and P.V. Harris, A possible functional role for a new class of eukaryotic DNA polymerases. Curr Biol, 1997. 7(12): p. R743-4.
- Burgers, P.M., et al., Eukaryotic DNA polymerases: proposal for a revised nomenclature. J Biol Chem, 2001. 276: p. 43487-43490.
- Garc¿a-G¿mez, S., et al., PrimPol, an archaic primase/polymerase operating in human cells. Mol Cell, 2013. 52(4): p. 541-53.
- Bianchi, J., et al., PrimPol bypasses UV photoproducts during eukaryotic chromosomal DNA replication. Molecular Cell, 2013. 52(4): p. 566-73.
- Iyer, L.M., et al., Origin and evolution of the archaeo-eukaryotic primase superfamily and related palm-domain proteins: structural insights and new members. Nucleic Acids Res, 2005. 33(12): p. 3875-96.
- Clark, K., et al., GenBank. Nucleic Acids Res, 2016. 44(D1): p. D67-72.
 BACK TO TOP
BACK TO TOP