Basic mechanism of TLS in eukaryotic cells
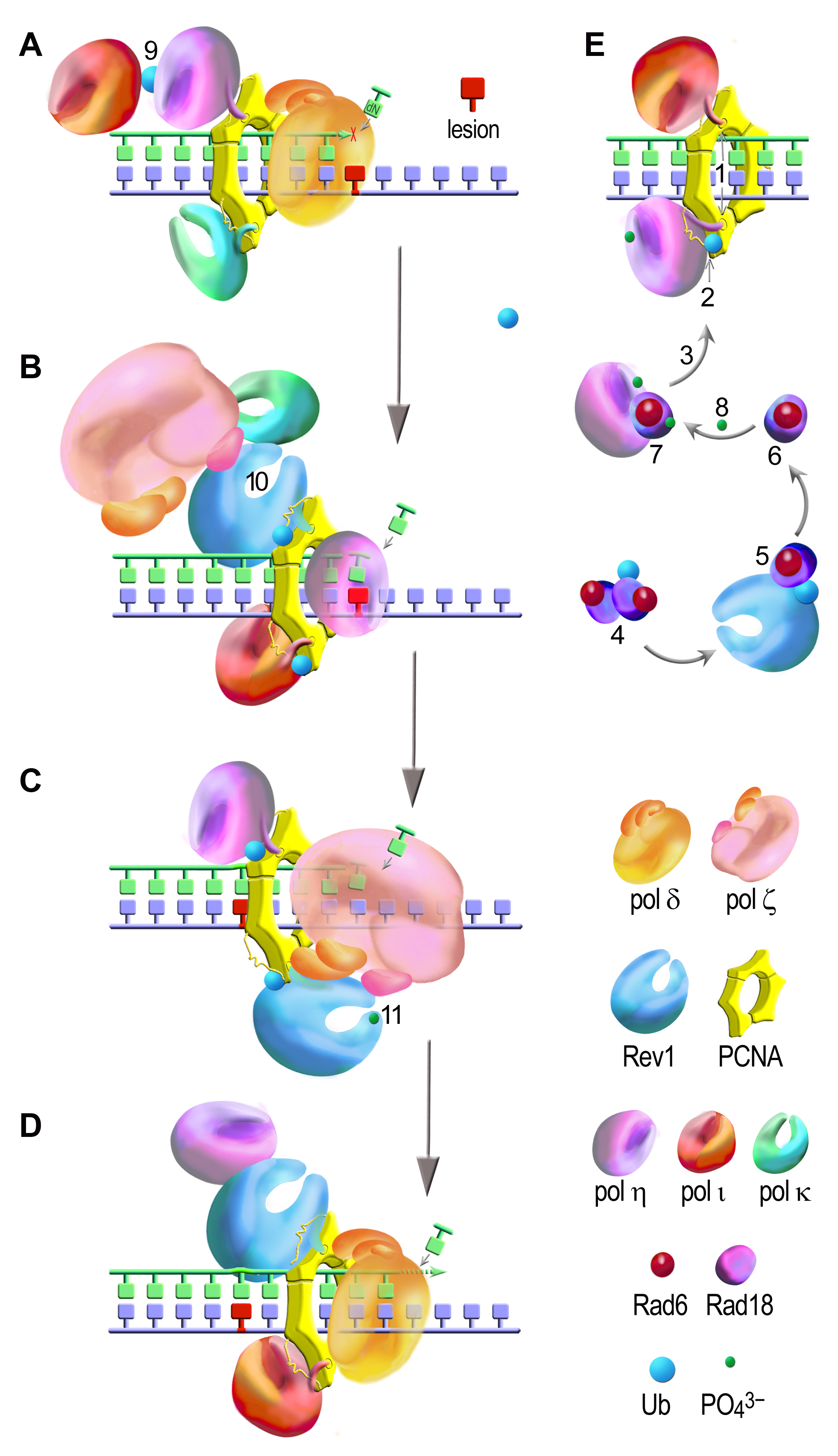
TLS is a multi-step process involving (A) stalling of the replicative DNA polymerase (here shown for pol δ consisting of PolD1, PolD2, PolD3, and PolD4 subunits) at damaged sites (red); (B) PCNA ubiquitination, recruitment of a TLS polymerase to the primer terminus (here shown for pol η) and nucleotide (mis)insertion opposite the lesion; (C) extension (catalyzed by pol ζ) of the resulting primer termini by several nucleotides; (D) deubiquitination of PCNA and release of a TLS polymerase after bypass completion allowing the normal replication restart. TLS is regulated through an elaborate network of protein-protein interactions and posttranslational modifications (shown for different polymerases in A-E; for the details dee Vaisman & Woodgate, Translesion DNA polymerases in eukaryotes: what makes them tick? Crit Rev Biochem Mol Biol, 2017). TLS polymerases are recruited to the replication factories via interactions with PCNA (E), which due to the trimeric structure and various binding sites, can accommodate multiple partners (A-E). The initial interactions through the PIP boxes (1) are too unstable, which makes them hard to be detected in replication foci. DNA damage reinforces the PCNA/TLS polymerases interactions due to monoubiquitination of PCNA (2) that is catalyzed by the RAD6/RAD18 complex (3). In normal cells, Rad18 forms a dimer consisting of a monoubiquitinated and nonubiquitinated proteins (4). Non-modified Rad18 (5) that is required for PCNA ubiquitination is released from the dimer via an interaction with Rev1 (6) and is stabilized by an interaction with polη (7). Damage-induced phosphorylation of pol η and Rad18 facilitates their interaction and localization to replication foci (8). PCNA ubiquitination is further promoted by the binding of the polymerase molecules via their UBD domains (2). Pol η has a higher affinity for ubiquitinated PCNA and this interaction is sufficiently stable for pol η to form detectable foci, while the accumulation of pol ι requires its interaction with pol η (9 in the panel A). Monoubiquitination of either polη or polι enhances the interaction between these two polymerases (9). Cells use another protein binding platform, Rev1, which serves as a “polymerase bridge” for the assembly of two TLS polymerases (shown in panel B for pol ζ (through Rev7 as shown here, and through the PolD3 subunit) and pol k (10)) and might facilitate the switch from the “inserter” (here shown for pol η) to the “extender” (pol ζ). Interactions between Rev3 (through Rev7 and PolD3) and Rev1 are possibly facilitated by Rev1 phosphorylation (11). Such interactions stabilize binding of pol ζ (through the PIP motif of PolD3) to ubiquitinated PCNA and thereby enhances pol ζ-dependent TLS.
 BACK TO TOP
BACK TO TOP