Basic mechanism of TLS in prokaryotic cells
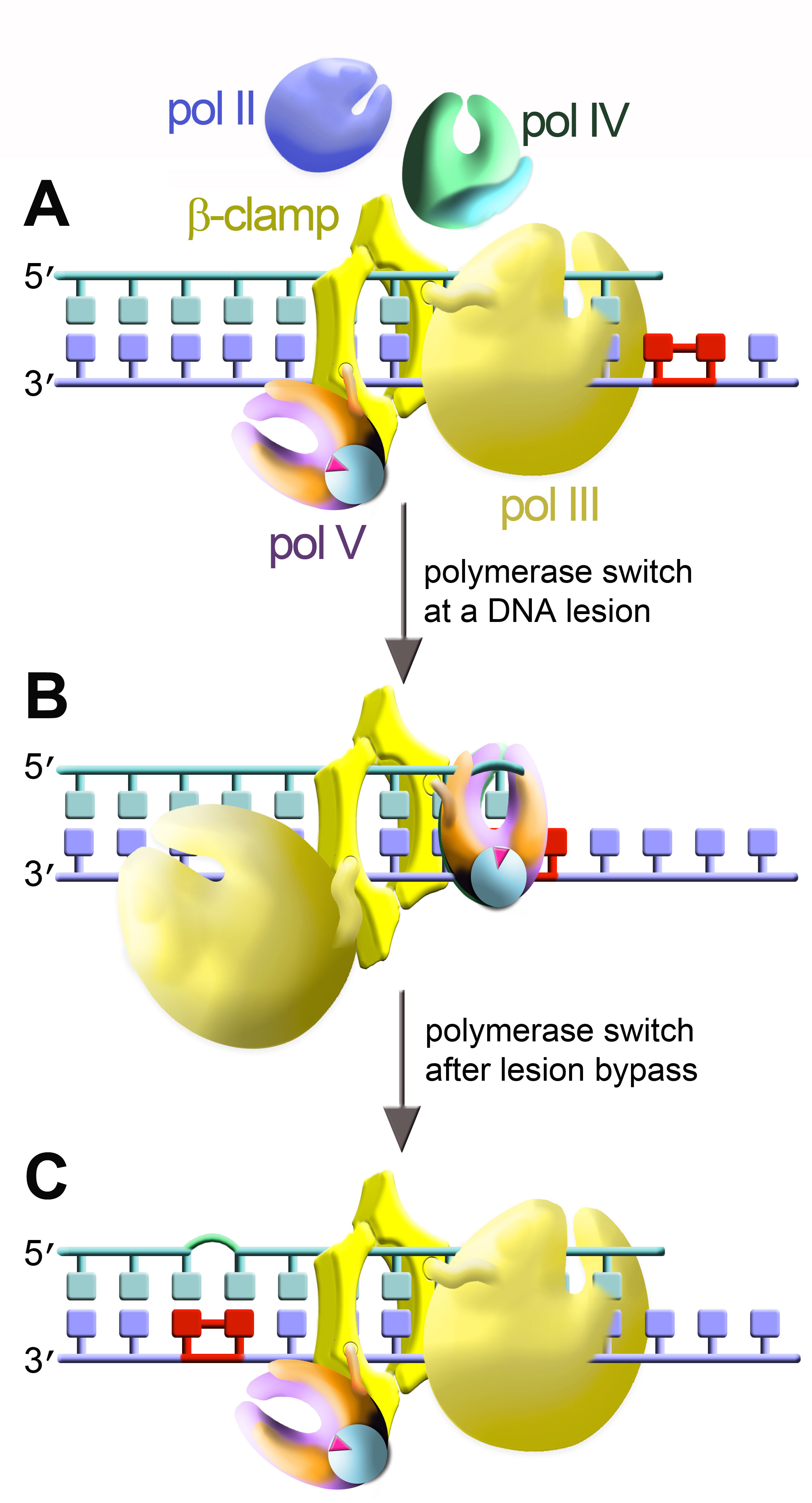
The cartoon depicts bacterial DNA polymerases: replicative holoenzyme pol III [yellow] and three SOS-inducible polymerases, pol II (blue), pol IV (green), and pol V (violet/orange).
Two DNA polymerases are shown simultaneously bound to the dimeric β-clamp.
(A) The main replicase, pol III, is engaged at the primer terminus. (B) At a DNA lesion, pol III disengages from the primer terminus and TLS polymerase rotates into position so as to promote TLS. In this cartoon TLS is catalyzed by pol V (UmuD'2C•RecA•ATP complex formed by the transfer of a single RecA protomer [light blue] along with ATP from the 3' tip of a RecA nucleoprotein filament to the heterotrimer composed of the catalytic UmuC subunit [violet] and a homodimer of UmuD' [orange]. (C) After TLS has occurred, pol V is replaced by pol III, which resumes genome duplication. This cartoon is based on the "tool belt" model for polymerase switching originally proposed by Pages and Fuchs (Pages, V., and R. P. Fuchs. 2002. How DNA lesions are turned into mutations within cells? Oncogene 21:8957–8966).
 BACK TO TOP
BACK TO TOP