Research
Meiotic pairing initiation
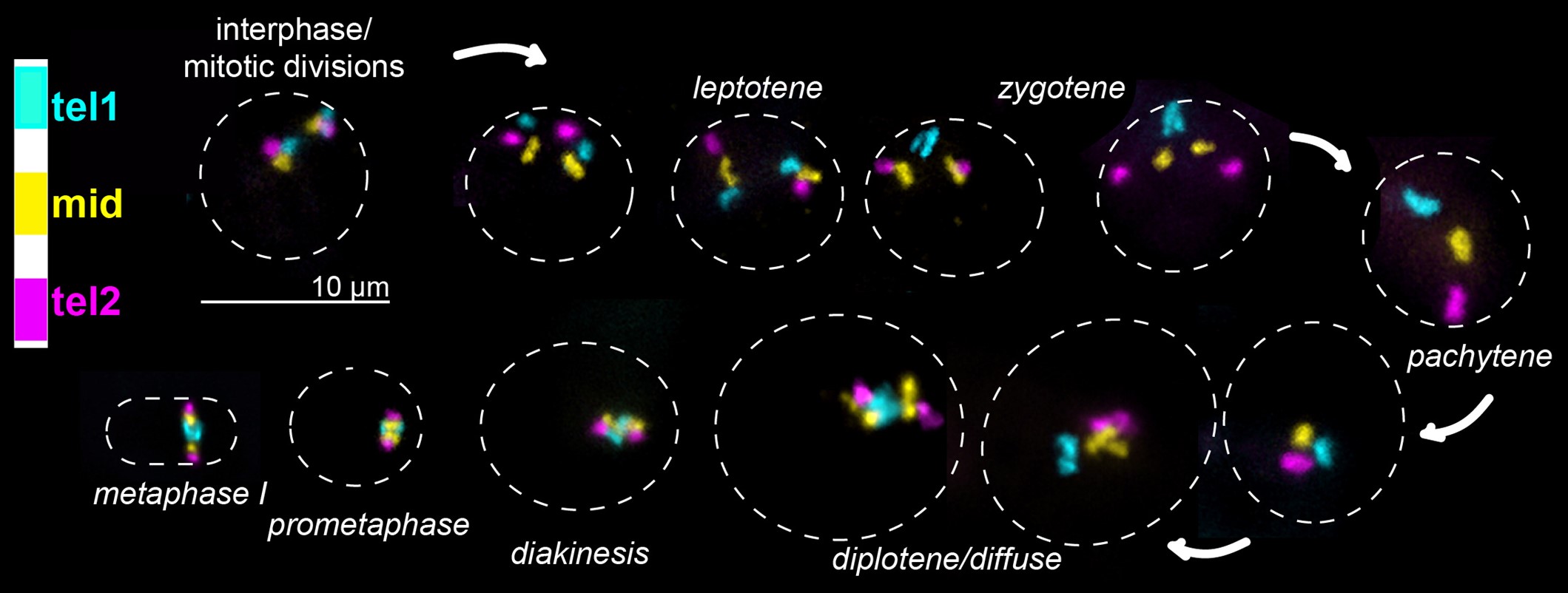
Precise homolog pairing is essential for accurate meiotic chromosome segregation in species from plants to humans. Failure to accurately segregate chromosomes in meiosis can lead to nondisjunction and infertility or aneuploidy in progeny. Yet, the mechanisms regulating homolog recognition and pairing are unknown. This has been particularly mysterious since the discovery that homolog recognition and alignment are independent of DNA double-strand breaks (DSBs) in most species. Instead, DSBs are required to form crossovers, which help stabilize pairing interactions after they are established.
We previously employed the highly specific Oligopaint fluorescence in situ hybridization (FISH) technology to interrogate homolog recognition and pairing initiation in the silkworm moth Bombyx mori (Rosin et al., 2021, PLOS Genetics 
One of our current research goals is to investigate the mechanisms behind this gene-rich-associated pairing and investigate whether transcription, the transcription machinery, or transcripts themselves facilitate homolog recognition and meiotic pairing initiation in moths and beyond.
Meiotic chromosome segregation
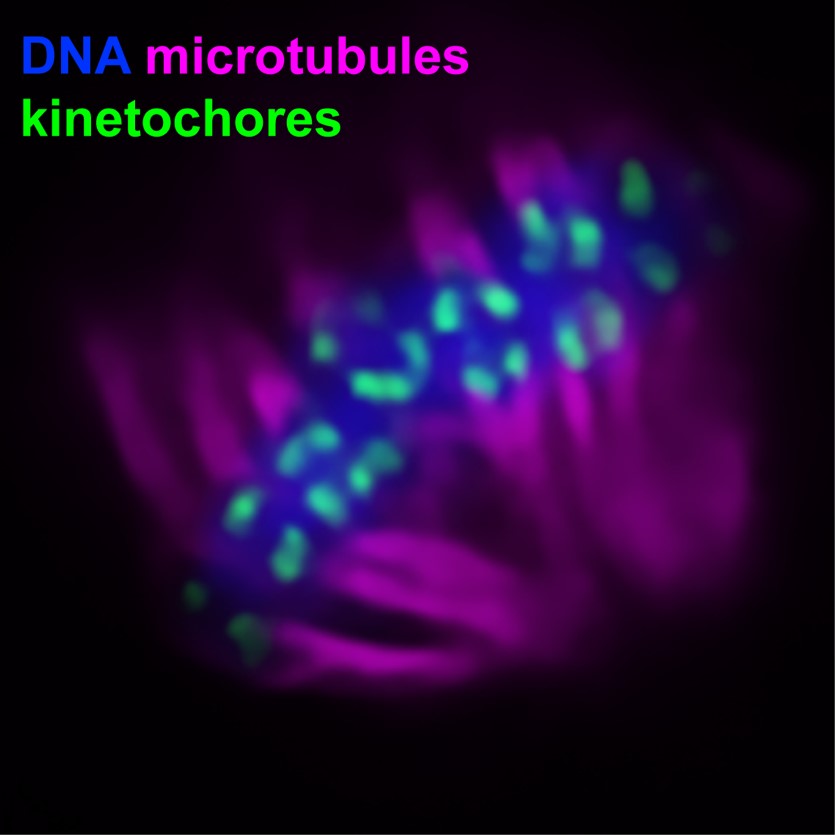 In eukaryotic species from yeast to humans, the segregation of chromosomes in mitosis and meiosis depends on the presence of a specialized histone H3 variant called CENP-A (centromere protein A). Interestingly, a recent study found that CENP-A has been lost in several insect lineages, including Lepidoptera (moths and butterflies). How these species segregate their chromosomes without CENP-A is an area of active study in our lab as well as others.
In eukaryotic species from yeast to humans, the segregation of chromosomes in mitosis and meiosis depends on the presence of a specialized histone H3 variant called CENP-A (centromere protein A). Interestingly, a recent study found that CENP-A has been lost in several insect lineages, including Lepidoptera (moths and butterflies). How these species segregate their chromosomes without CENP-A is an area of active study in our lab as well as others.
Our previous work found that these species harbor rather canonical kinetochores during meiosis despite not having CENP-A (Rosin et al., 2021, PLOS Genetics 
Condensin II function and genome stability
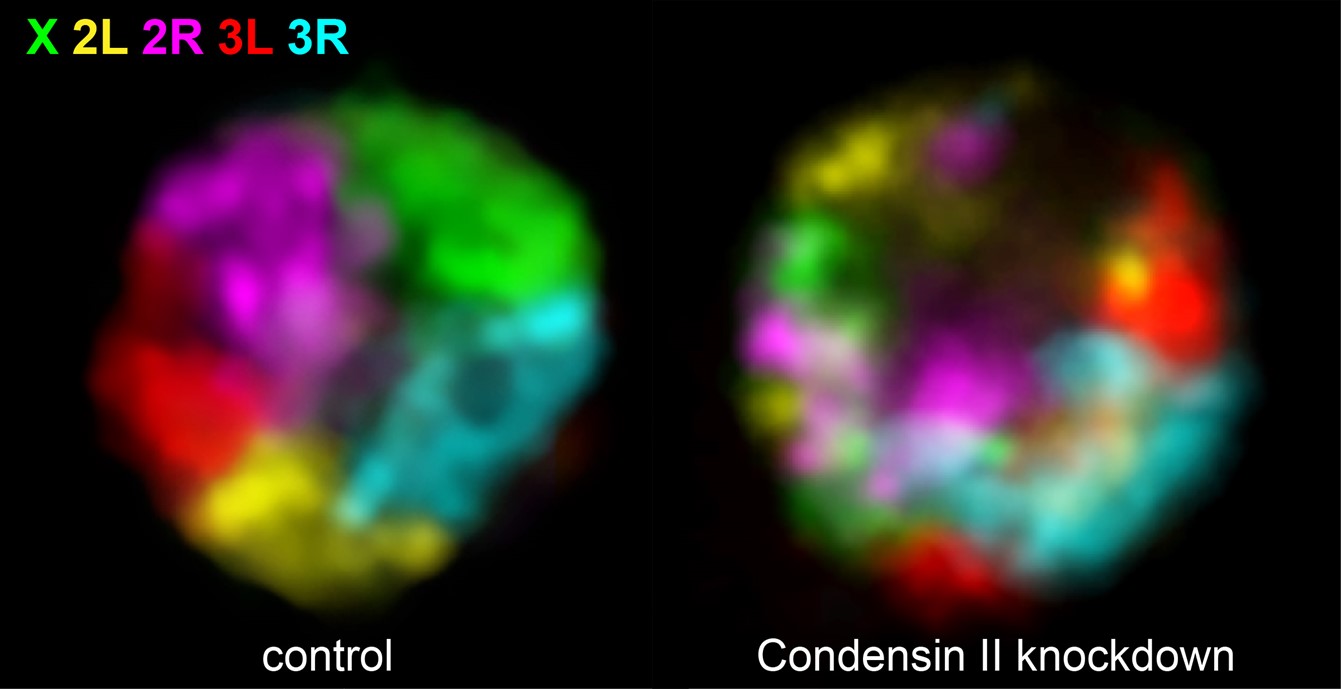
Condensins and related proteins are ring-shaped architectural proteins that are highly conserved and found from bacteria to humans. They are known for their roles in mitotic genome compaction (Condensin I and Condensin II) and sister chromatid cohesion (Cohesin). Condensin II also plays essential roles in interphase genome organization and genome stability (Rosin et al., 2018, PLOS Genetics 

An unexpected role of Condensin II is that it antagonizes interactions between homologous chromosomes (maternal and paternal copies) in somatic cells in Drosophila. Our lab is investigating whether this function of Condensin II is conserved across species and furthermore, we are interrogating potential roles of Condensin II in the regulation of meiotic homolog pairing.
Model Systems
Insects: Bombyx mori, Plodia interpunctella, Drosophila melanogaster
Mammals: Mus musculus

Why use moth model systems in biomedical research?
Lepidopteran insects (moths and butterflies; Leps) are ideal model systems to study chromosome dynamics and address long-standing, fundamental questions in genome and reproductive biology.
In addition to being important industrial and agricultural insects, Leps are excellent model systems for biomedical research, especially for the study of homolog pairing and meiosis. The problem of the search for homology in cells becomes increasingly complicated as chromosome number increases. While pioneering studies in Drosophila and C. elegans have provided many answers about pairing regulation, these species harbor only 4 and 6 chromosomes, respectively. Like these classical model systems, Leps are genetically tractable and easy to rear in a laboratory setting. However, unlike flies and nematodes, Leps typically have 28-32 chromosomes, making genome dynamics and the search for homology more similar between Leps and mammals. However, unlike mammalian models, Leps have a short life cycle and are inexpensive to maintain.
Importantly, the recent sequencing of many Lep genomes has revealed a high degree of sequence homology between Lep genes and mammalian disease genes. Finally, Leps are large for insects, providing more tissue for genomics-based approaches. Even at the cellular level, Lep germline cells are much larger than fly germline cells, allowing for high resolution imaging without the need for expansion technology.
Tools
Oligopaints
One of the main approaches used in our lab is Oligopaint DNA FISH (originally developed in Ting Wu’s lab at Harvard). Unlike BAC-based FISH probes, Oligopaints are computationally designed probes based on genomic sequence. This allows for only unique, single copy sequences to be labeled, significantly increasing the specificity and resolution of FISH. The flexibility and scalability of the Oligopaint design pipeline can be leveraged to generate chromosome paints for any species with a sequenced genome. These paints can then be used to image chromosomes or sub-chromosomal regions in any tissue type, cell cycle stage, or developmental stage of interest to address fundamental questions of chromosome organization and cell biology.
More information about Oligopaints can be found at the following sites:
Live Imaging
In addition to fixed imaging techniques, our lab employs live cell imaging approaches. While the snapshots obtained by imaging fixed cells can provide a lot of information, live imaging allows cell dynamics to be captured in four dimensions.
Genomics
In addition to imaging-based approaches, our lab takes advantage of genomics technologies such as Chromatin immunoprecipition (ChIP)-seq, RNA-seq, Assay for Transposase-Accessible Chromatin (ATAC)-seq, and High-throughput Chromosome Conformation Capture (Hi-C). Together, these approaches allow us to map architectural protein binding sites throughout the genome and directly correlate this with 3D chromatin organization and gene expression. By combining these tools with the single-cell nature of microscopy, we are able to gather a more complete picture of chromatin organization and function.
 BACK TO TOP
BACK TO TOP