Research
All tissues and organs in adult animals arise from a single cell: the zygote. We use zebrafish embryos as a research system to explore how zygotes routinely produce the tissues needed in healthy adults. These vertebrate embryos develop quickly and are transparent and externally fertilized, facilitating microscopy-based research.

To create the many types of tissues required in a healthy adult, the developing embryo must activate genes that specify different tissues in stereotyped regions. In other words, embryos must reliably produce diverse patterns of gene expression to create functional adults. Perturbed gene expression during development can lead to congenital defects. Our primary goal is to understand how diverse gene expression patterns arise reproducibly in developing tissues.
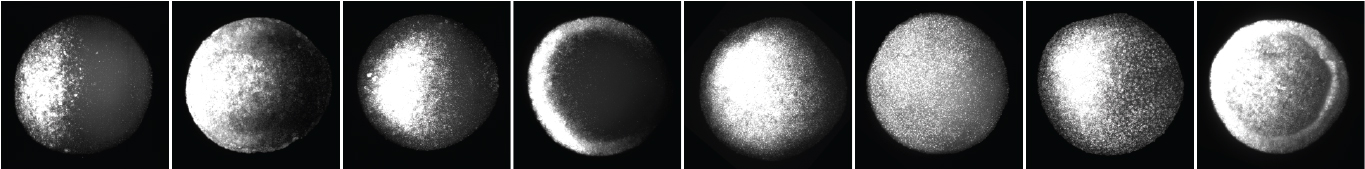
Much of the gene expression in embryos is controlled by signaling molecules that spread through tissues and interact with cells to activate genes. We seek to understand how the spread of signaling molecules is regulated, because the distribution of signaling molecules contributes to resulting patterns of gene expression. The in vivo biophysical properties of signaling molecules, such as their mobility and stability, influence their distribution. We use in vivo methods including FRAP and FDAP to measure signaling molecule mobility and stability, respectively, to understand how these properties are regulated and how they affect distribution.
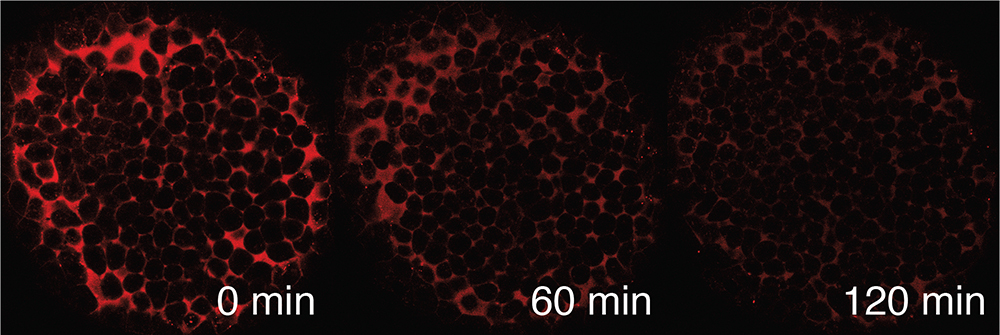
As signaling molecules spread through embryonic tissues, they interact with cells and activate gene expression. Genes can have unique responses to signaling that contribute to differences in their expression patterns. We seek to understand what features of signaling are “read out” by genes: how do signaling levels, duration, and dynamics affect the expression of different genes? In addition, multiple different kinds of signaling molecules are usually present in developing tissues. How are responses to one type of signaling molecule affected by the presence of a second (or third)? To understand how diverse gene expression patterns arise in developing tissues, we are working towards a more comprehensive understanding of the input/output relationship between signaling and gene expression in vertebrate embryos.
One of the methods we use to explore these questions is molecular optogenetics. These approaches allow researchers to manipulate biological processes using light by coupling light-responsive proteins, often from plants or bacteria, to different biological effectors. For example, it is possible to experimentally manipulate protein localization, gene expression, cell mobility, and signaling using light.
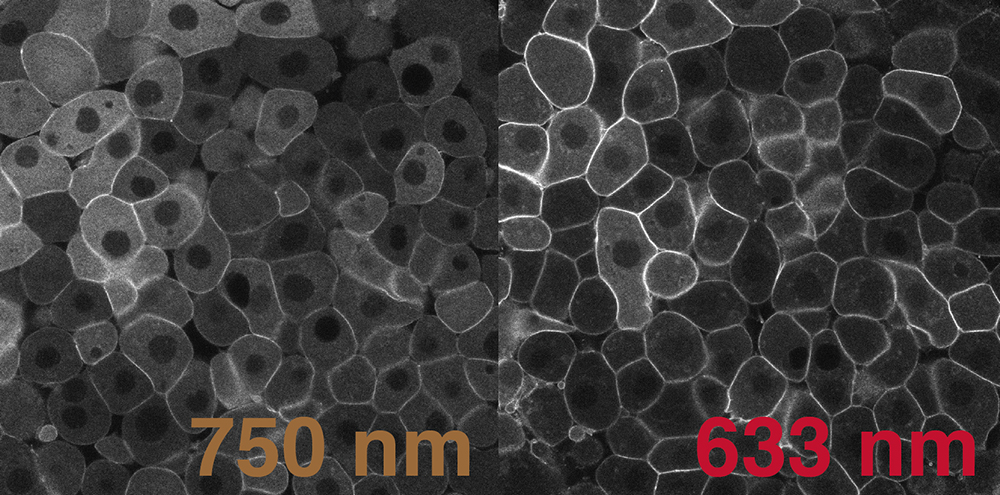
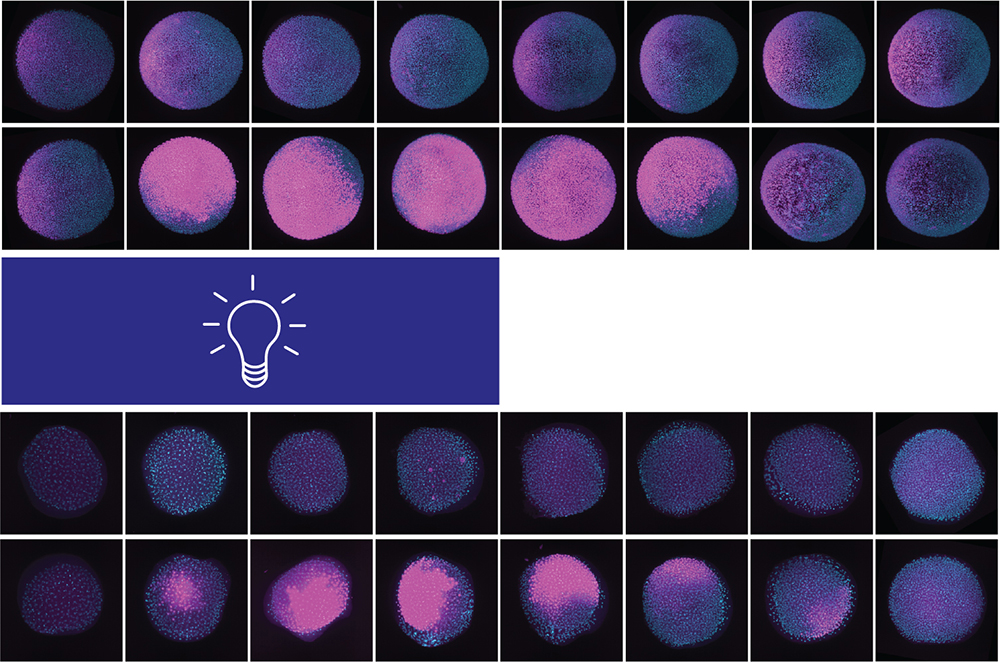
Molecular optogenetics has important advantages compared to conventional pharmacological or genetic methods: these approaches are often reversible, fast, and tunable, and can offer a high degree of spatiotemporal control. Because it is possible to control signaling levels, duration, and dynamics by adjusting light exposure, we can assess the input/output relationship between signaling and gene expression in vivo. We are also exploring approaches to simultaneously activate several signaling pathways with different wavelengths of light, providing an ideal platform to study the interpretation of multiplexed signaling.
 BACK TO TOP
BACK TO TOP