Research
Targeting nucleo-cytoplasmic transport proteins for selective degradation
NPCs consist of multiple copies of roughly 30 proteins (nucleoporins), which form a series of stable sub-complexes. Many nucleoporins play additional roles during mitosis, and some sub-complexes localize to kinetochores, where they facilitate chromosome segregation. Understanding the activities of individual nucleoporins in each of these contexts has been complicated by their multi-faceted nature: for example, RNAi (RNA interference)–mediated depletion of nucleoporins in mammalian cells causes phenotypic defects, including altered gene expression and abnormal mitotic progression. However, the abundance and unusual stability of these proteins requires an extended interval for their depletion, so that many phenotypes could be indirect consequences of disrupted nuclear trafficking as the number and quality of NPCs decline.
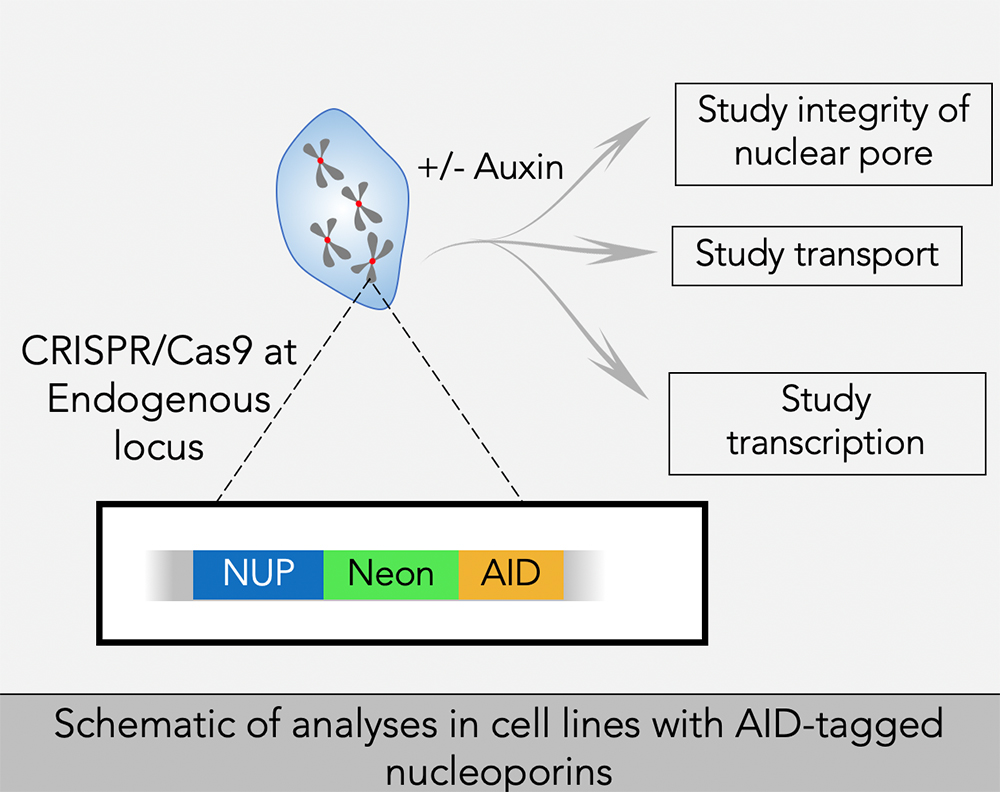
To better address the role of individual nucleoporins, we adapted AID strategies for selective and rapid degradation of individual proteins. We are using CRISPR/Cas9 to construct cell lines in which sequences encoding AIDs are inserted into both alleles of targeted genes of human tissue-culture cells that stably express the Transport Inhibitor Response 1 (TIR1) protein. TIR1 acts as a subunit of the SCF ubiquitin ligase complex, so that the AID–tagged fusion proteins undergo rapid, selective degradation upon addition of the plant hormone auxin. In general, we also add a fluorescent tag to the targeted proteins, allowing the degradation to be monitored visually as well as biochemically.
We have been successful in developing lines that allow conditional depletions of nucleoporins associated with the cytoplasmic and nuclear faces of the NPC, as well as nucleoporins that reside within the central domain of the NPC. We are examining the function of these nucleoporins in interphase and mitosis, as discussed below. We are continuing our efforts to make lines for conditional depletion of all remaining nucleoporins, as well as collaborating extensively with groups who have developed approaches to address other issues of nucleoporin function.
Using these cell lines, we will study the role of nucleoporins in nuclear organization and function, mitosis, nuclear transport, transcription and overall nuclear architecture.
Mitotic regulation of the Ran GTPase
Ran is a Ras–family GTPase that plays critical roles in many cellular processes, including nucleo-cytoplasmic transport, nuclear envelope assembly, and mitotic spindle assembly. Ran alternates between GDP– and GTP–bound forms. In interphase cells, GTP–bound Ran (Ran–GTP) is the major form in the nucleus, while GDP–bound Ran (Ran-GDP) is the predominant form in the cytoplasm. The asymmetrical distribution of Ran–GTP and Ran-GDP drives cargo transport between the nucleus and cytoplasm through karyopherins, a family of nuclear transport carrier proteins that bind to Ran–GTP. During mitosis, after nuclear envelope breakdown, Ran–GTP is concentrated in the region close to mitotic chromatin, while Ran-GDP is the major form distal to chromatin. The Ran–GTP gradient guides mitotic spindle assembly by releasing spindle assembly factors (SAFs) from karyopherins based on local Ran-GTP concentrations. In cells, the conversion of Ran–GDP to Ran–GTP is catalyzed by a Ran–specific guanine exchange factor called RCC1 (regulator of chromosome condensation 1) in vertebrates. The capacity of RCC1 to bind to chromatin establishes the asymmetrical distribution of Ran–GTP in interphase as well as the chromatin-centered Ran–GTP gradient in mitosis. Interestingly, RCC1’s association with chromatin is not static during the cell cycle and, in vertebrate systems, is regulated in a particularly dramatic fashion during anaphase. The regulation has not been correlated with post-translational modifications of RCC1, and the underlying molecular mechanism has not been reported.
RanBP1 is a highly conserved Ran–GTP–binding protein, which acts as co-activator of RanGAP1 (Ran GTPase–Activating Protein 1) and can form a heterotrimeric complex with Ran and RCC1 in vitro. We found that RCC1 not associated with chromosomes during mitosis is sequestered and inhibited in RCC1/Ran/RanBP1 heterotrimeric complexes and that the sequestration is crucial for normal mitotic spindle assembly. In addition, RanBP1 complex formation competes with chromatin binding to regulate the distribution of RCC1 between the chromatin-associated and soluble fractions. Moreover, we identified a cell cycle–dependent phosphorylation on RanBP1 that modulates RCC1/Ran/RanBP1 heterotrimeric complex assembly and releases RCC1 to bind to chromatin; the phosphorylation is directly responsible for controlling RCC1 dynamics during anaphase. Together, our findings demonstrate novel roles of RanBP1 in spindle assembly and RCC1 regulation in mitosis. We are currently extending these findings to determine whether RanBP1 plays an analogous role in mammalian cells during mitosis.
SUMO–family small ubiquitin-like modifiers in higher eukaryotes
SUMOs are ubiquitin-like proteins (Ubls) that become conjugated to substrates through a pathway that is biochemically similar to ubiquitination. SUMOylation is involved in many cellular processes, including DNA metabolism, gene expression, and cell-cycle progression. Vertebrate cells express three major SUMO paralogs (SUMO-1–3): mature SUMO-2 and SUMO-3 are 95% identical, while SUMO-1 is 45% identical to SUMO-2 or SUMO-3 (where they are functionally indistinguishable, we collectively call SUMO-2 and SUMO-3 SUMO-2/3). Like ubiquitin, SUMO-2/3 can be assembled into polymeric chains through the sequential conjugation of SUMOs to each other. Many SUMOylation substrates have been identified. SUMOylation promotes a variety of fates for individual targets, dependent upon the protein itself, the conjugated paralog, and whether the conjugated species contains a single SUMO or SUMO chains.
SUMOylation is dynamic owing to rapid turnover of conjugated species by SUMO proteases. Both post-translational processing of SUMO polypeptides and deSUMOylation are mediated by the same family of proteases, which play a pivotal role in determining the spectrum of SUMOylated species. This group of proteases is called Ubl–specific proteases (Ulp) in yeast and Sentrin-specific proteases (SENP) in vertebrates. There are two yeast Ulps (Ulp1p and Ulp2p/Smt4p) and six mammalian SENPs (SENP1, SENP2, SENP3, SENP5, SENP6, and SENP7). SENP1, SENP2, SENP3, and SENP5 form a Ulp1p–related sub-family, while SENP6 and SENP7 are more closely related to Ulp2p. Yeast Ulps have important roles in mitotic progression and chromosome segregation. We defined the enzymatic specificity of the vertebrate SENP proteins and analyzed their key biological roles.
Ulp1p localizes to NPCs, is encoded by an essential gene, and is important for SUMO processing, nucleocytoplasmic trafficking, and late steps in the ribosome biogenesis pathway. Humans possess two NPC–associated SENPs: SENP1 and SENP2. While SENP2 is dispensable for cell division, mammalian SENP1 was recently shown to play an essential role in mitotic progression. We are currently analyzing AID–tagged alleles of both SENP1 and SENP2 to assess their roles in both interphase (nuclear trafficking and gene expression) and during mitosis (kinetochore function and mitotic progression), as well as their dependence upon individual nucleoporins for their targeting to the interphase NPC.
The role of the IRBIT protein in tissue homeostasis
We recently reported a conserved role for the IRBIT protein (IP3-receptor-binding protein released with inositol 1,4,5-trisphosphate) in inhibiting ribonucleotide reductase (RNR), an enzyme that produces deoxynucleotide triphosphates (dNTPs) within the cell for DNA synthesis. We further found that mammalian tissue-culture cells show altered cell-cycle progression and potentially disrupted genome stability in the absence of IRBIT. In the same report, we showed that this mechanism is conserved between humans and flies (Drosophila melanogaster). Therefore, in collaboration with Mihaela Serpe and Brian Oliver, we chose to use flies as a model organism to understand the role of this mechanism in development and tissue homeostasis.
In situ hybridization shows IRBIT expression in regions destined to become the midgut during embryogenesis, and IRBIT is highly expressed in the adult midgut. The Drosophila midgut has a tubular structure and is surrounded by visceral muscles. The adult midgut possesses a monolayered epithelium that is composed of four distinct cell types : intestinal stem cells (ISCs), undifferentiated progenitor cells called enteroblasts (EBs), specialized absorptive enterocytes (ECs), and secretory enteroendocrine cells (EEs). The midgut is maintained through division of ISCs, giving rise to EBs, which in turn differentiate into EEs. Nutrients are absorbed from the lumen of the gut, which also contains a complex microbiota; the midgut acts both as a niche for commensal microbes and as the first line of defense for against microbial pathogens. Like the intestine of vertebrates, the epithelium of the midgut has a remarkable regenerative capacity, which has been extensively exploited for the study of stem cell–driven tissue self-renewal, as well as tissue homeostasis during aging.
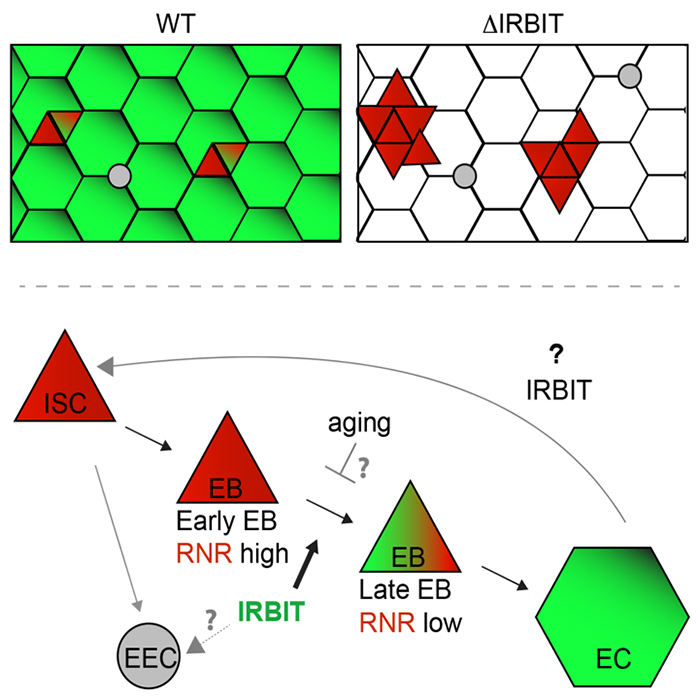
We examined IRBIT’s potential role in the midgut by generating an IRBIT null fly (IRBIT–/–). The midguts of one-day-old wild-type and IRBIT–/– flies were essentially indistinguishable at the tissue-architecture level. However, we observed a rapid loss of tissue homeostasis in the IRBIT–/– flies, with a progressive increase in relative numbers of undifferentiated enteroblast progenitor cells and tissue dysplasia. IRBIT–/– flies also show fewer cell-cell contacts when stained for junctional proteins in the posterior midgut epithelium and altered gene expression patterns reminiscent of changes associate with inflammation and aging. The phenotypes are fully rescued through expression of full-length IRBIT, and further experiments suggested that altered dNTP pools likely contribute to the IRBIT–/– phenotypes.
Together, our data suggest that IRBIT plays a key role in tissue homeostasis in the fly midgut by antagonizing RNR in nucleotide metabolism. We are currently examining the interplay of IRBIT with inflammatory pathways and microbiota changes that are characteristic features of aging in flies.
 BACK TO TOP
BACK TO TOP