Research
Integral membrane enzymes that catalyze protein lipidation
Structure and Mechanism of protein lipidation by DHHC S-acyltransferases
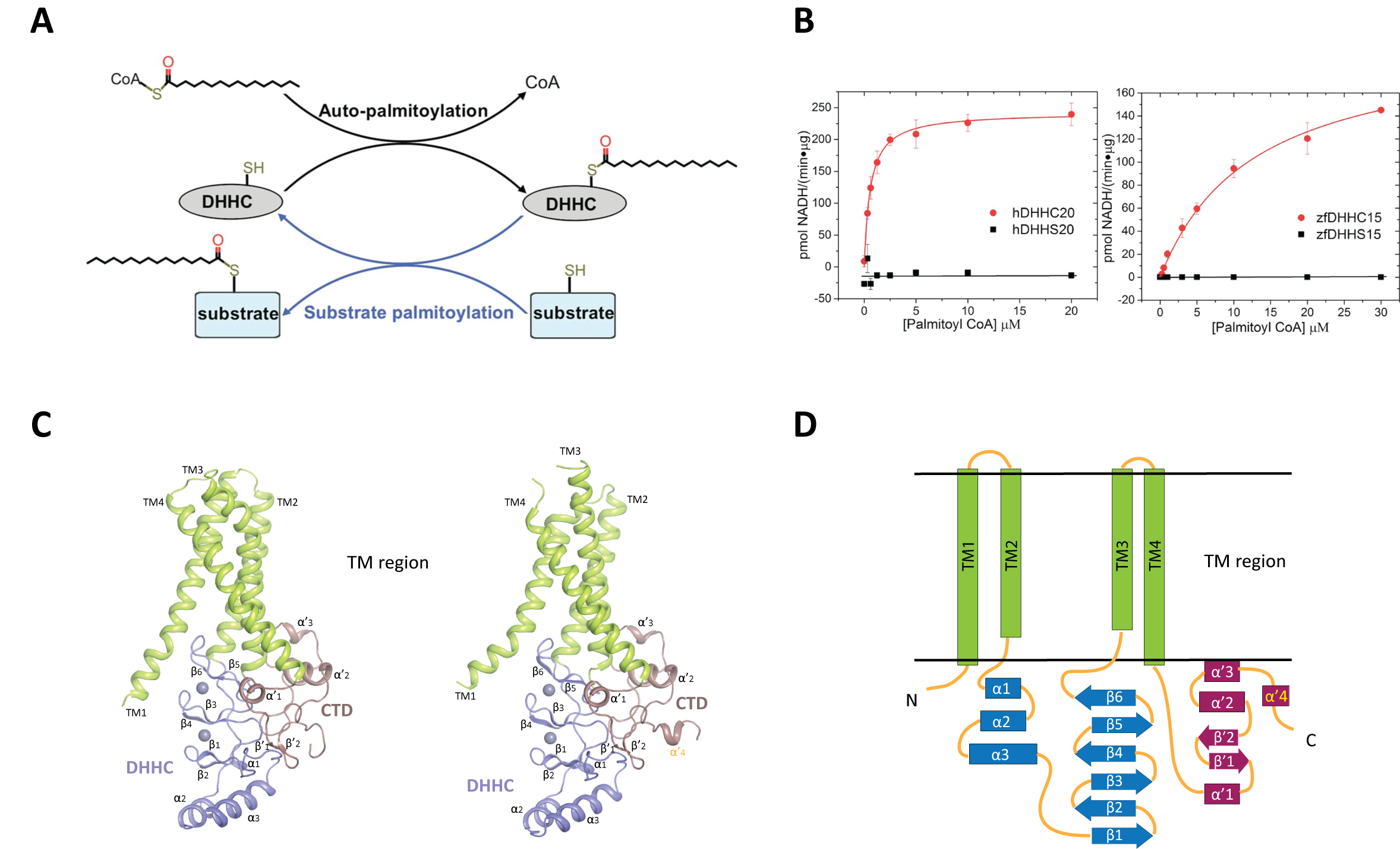
Covalent attachment of lipids is one of the most prevalent forms of post translational protein modification. Although physico-chemically it amounts to attachment of a mere “hydrophobic stick”, protein lipidation can modulate protein function by altering structure, folding, interaction with other proteins, degradation and importantly, attachment to membranes. Of the different forms of protein lipidation, the most pervasive form is attachment of a fatty acid to internal cysteines through a thioester. This is known as protein S-acylation or more commonly, protein palmitoylation, owing to the prevalence of the attachment of the 16 carbon palmitoyl moiety through this kind of modification. In the cellular milieu, thioesters can be cleaved by thioesterases and thus protein S-acylation is unique in being the only reversible form of protein lipidation. Protein palmitoylation is catalyzed by members of the DHHC family of eukaryotic integral membrane enzymes so called because of the catalytic Asp-His-His-Cys motif in a cysteine rich domain (CRD) that resides in an intracellular loop. There are 23 DHHC enzymes in humans and several of them have been linked to a number of diseases, especially neuropsychiatric diseases and cancer.
We are interested in the molecular mechanism of DHHC palmitoyltransferases. Very recently we published the first crystal structures of two members of this family (Science, 2018, 359, eaao6326), together with the structure of one of them covalently conjugated to an irreversible inhibitor. These structures have opened up a number of interesting questions that we are currently investigating together with other members in the family.
Click image to enlarge.
Structure and mechanism of Porcupine, an essential enzyme in the Wnt signaling pathway
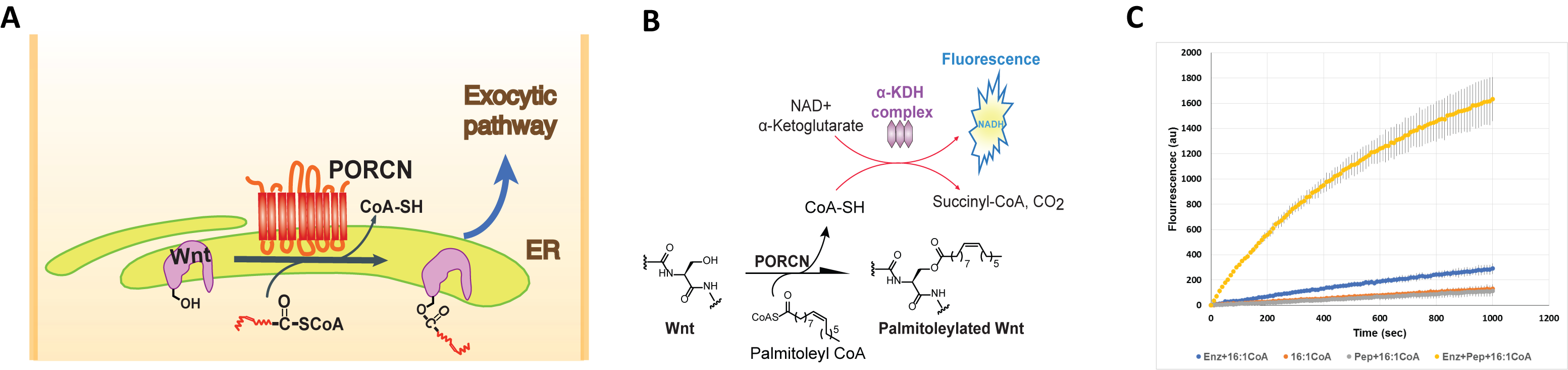
Three members of the membrane bound O-acyltransferase (MBOAT) family, a distinct family of integral membrane enzymes, also catalyze protein lipidation. What sets these three apart is that each of these three enzymes, Goat, Hhat and Porcupine, has a unique substrate, all secreted signaling proteins and each uses a distinct fatty acyl substrate. We are interested in the structure and mechanism of Porcupine, an essential enzyme for the biogenesis and secretion of Wnt. Porcupine is an ER resident transmembrane enzyme and modifies Wnt with an unsaturated fatty acid, the 16 carbon palmitoleic acid. Despite years of genetic and cell-based studies suggesting that Porcupine is the acyltransferase for Wnt, there was no report of biochemical purification and in vitro reconstitution to demonstrate this hypothesis. We reported (JBC, 2019, 294, 231) the first successful purification of human Porcupine and showed with an in vitro assay that it acylates Wnt. Fatty acyl selectivity of Porcupine suggested that the fatty acyl substrate binds to a kinked cavity in Porcupine matching the ground state geometry of the unsaturated acyl substrate.
Click image to enlarge.
Molecular mechanisms of iron transport and trafficking
Iron is an essential micronutrient for life on Earth due to its redox properties and abundance. Besides many iron containing proteins that contain iron-binding sites, iron is also assembled into cofactors like heme and iron-sulfur clusters. These components are then inserted into enzymes and other proteins for use in a wide range of crucial cellular activities like electron transport in respiratory chain complexes, regulatory sensing, photosynthesis, and DNA repair.
Structure and mechanism of Mitoferrin, the only known major transporter of iron into mitochondria
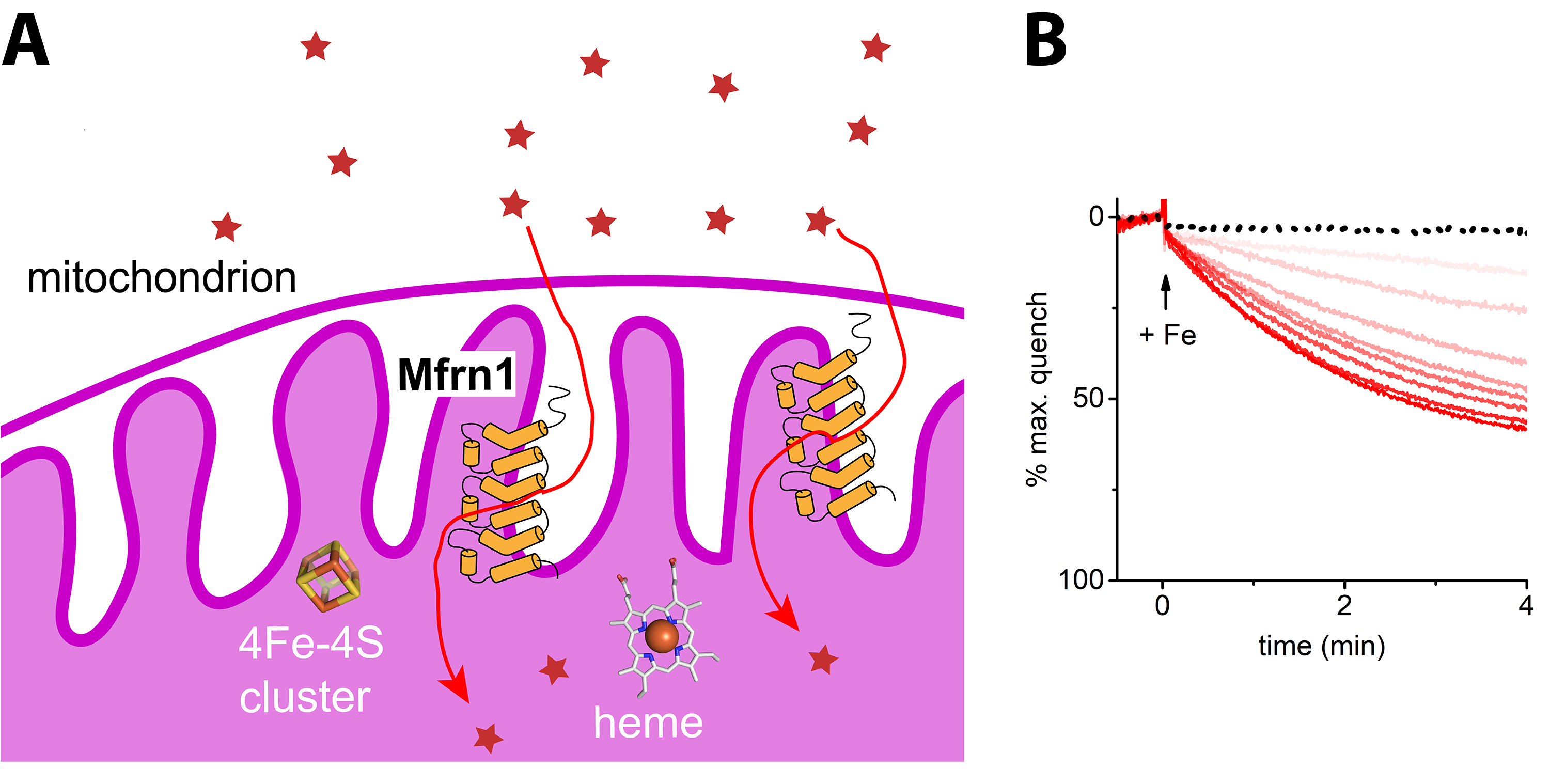
Mitochondria are not only the cellular power plants, they are also manufacturing centers for heme and iron-sulfur clusters. Genetic studies pointed toward a pair of mitochondrial carriers—termed Mitoferrins -1 and -2 in humans—as high-affinity iron importers. Although implicated as such, it had never been shown with an in vitro reconstitution assay with purified protein that Mitoferrin-1 and Mitoferrin-2 can indeed transport iron. We set up the first robust in vitro iron reconstitution assay, as part of our efforts to study Mitoferrin-1 structure and function and demonstrated unambiguously (JBC, 2018, 293, 3819) that Mitoferrin-1 transports iron(II) ions for supplying the metal to downstream targets in the mitochondrial matrix. We also found that Mitoferrin-1 is promiscuous for transporting first-row transition metals, implying the carrier can, in principle, supply manganese, zinc, and copper for essential matrix enzymes.
Click image to enlarge.
Hijacking of iron at the host-pathogen interface of Legionella pneuomophila by MavN, a highly conserved transporter
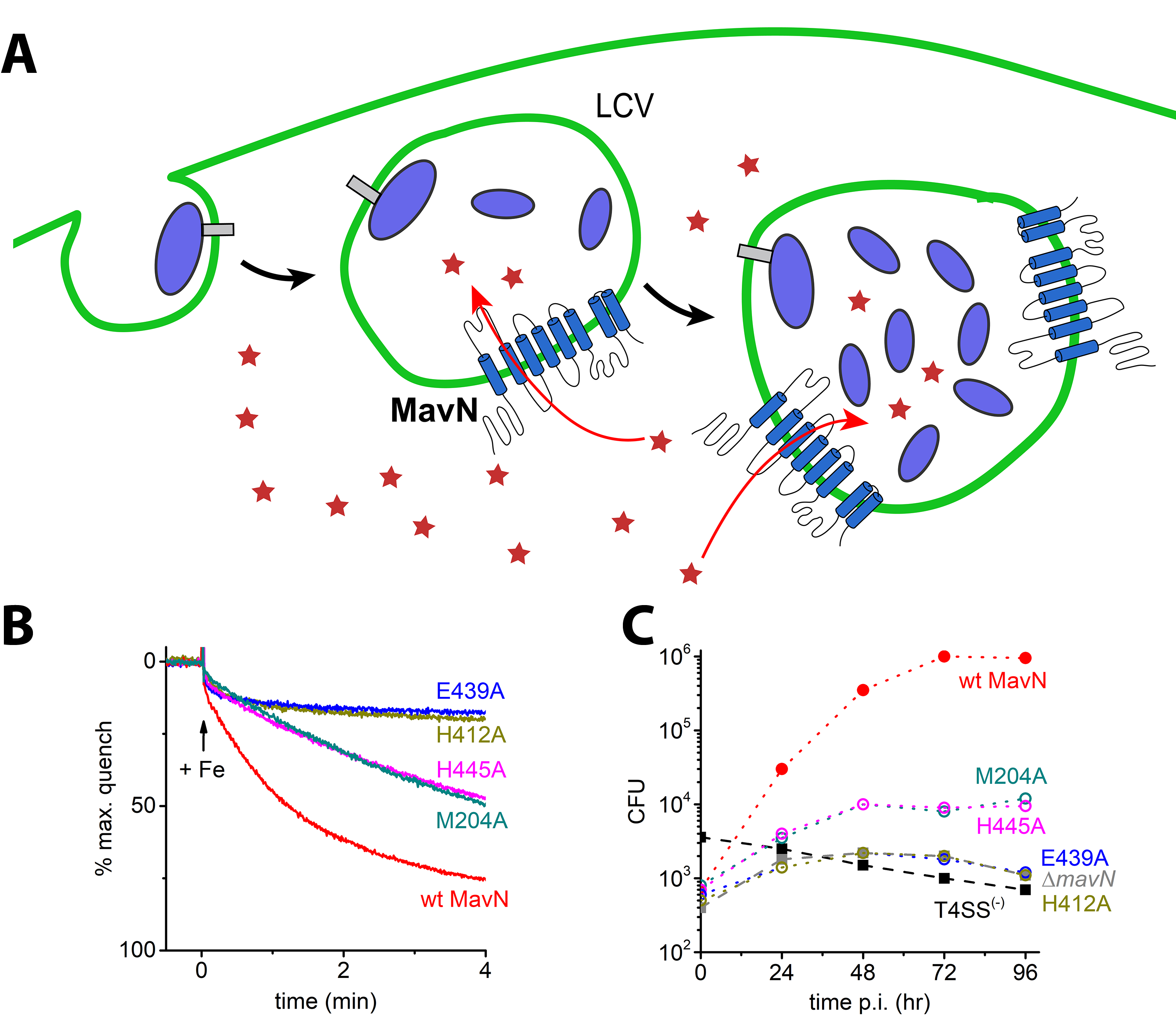
Iron is also a crucial nutrient for pathogens as well and starving them of metals is a central prong of the nutritional immunity response. Intracellular pathogens, such as Legionella pneumophila, have evolved sophisticated machineries to avoid immune system recognition while pilfering supplies from their hosts. In Legionella, the bacteria use an absolutely conserved effector protein, MavN, to support their proliferation inside host cells. Iron deficiency strongly induces MavN expression by Legionella and MavN was thus proposed to be an iron transporter. We recently demonstrated (PNAS, 2019,116,17775), using the same assay as we developed for Mitoferrin-1, that MavN can indeed transport iron in vitro and uncovered that Legionella proliferation is intimately tied to iron supplied through MavN.
Click image to enlarge.
Figure 4. (A) Legionella pneumophila is an intracellular pathogen that sequesters itself in an intracellular compartment called Legionella Containing Vacuole (LCV) and uses it as a site for proliferation and growth. MavN is a transmembrane protein that is synthesized in Legionella and inserted into the LCV membrane and is used for hijacking of iron from the host by Legionella. (B) Reconstitution of iron transport by wild type MavN and selected mutants using purified protein with a fluorescence-based assay in proteoliposomes. (C) Cell-based growth assay of MavN knockout Legionella with wild-type or mutants of MavN supplemented from a plasmid. There is a striking similarity between the in vitro transport assay and the in cellulo growth assay.
We are presently exploring the mechanistic and structural bases of substrate recognition and transport by Mitoferrin-1 and MavN how they interact with other chaperone proteins for metal trafficking.
 BACK TO TOP
BACK TO TOP