Research
 Studying gene regulation is critical to understanding how disruptions in developmental gene expression lead to birth defects and enable earlier diagnosis and potential intervention strategies. We investigate how genome folding not only fits the long DNA fiber within the constraints of the nuclear space but also is used as a critical mechanism of gene regulation during cell fate specification. This is a key fundamental question because transcription of many genes required for healthy development is controlled by non-coding regulatory elements—known as enhancers—that can be found at large genomic distances from their target genes. Our long-term scientific goal is to decipher how within a crowded nucleus, distal enhancers are guided specifically to their target genes and which mechanisms prevent off-target activation.
Studying gene regulation is critical to understanding how disruptions in developmental gene expression lead to birth defects and enable earlier diagnosis and potential intervention strategies. We investigate how genome folding not only fits the long DNA fiber within the constraints of the nuclear space but also is used as a critical mechanism of gene regulation during cell fate specification. This is a key fundamental question because transcription of many genes required for healthy development is controlled by non-coding regulatory elements—known as enhancers—that can be found at large genomic distances from their target genes. Our long-term scientific goal is to decipher how within a crowded nucleus, distal enhancers are guided specifically to their target genes and which mechanisms prevent off-target activation.
Identify and characterize DNA regulatory elements that facilitate or restrict long-distance activation.
Such elements may shape genome folding globally and indirectly modulate enhancer interactions—for example DNA Motifs bound by CTCF. Alternatively, some sequences may directly tether contacts with enhancers—CpG islands have been proposed to do this in vertebrates. We use unbiased and candidate-based functional approaches to discover novel elements and characterize their function.
Discover regulatory complexes that shape enhancer–promoter contacts
Identifying motifs will be the first step to pinpoint the regulatory complexes that establish enhancer contacts and how they induce transcription. We will focus on transcription factors as well as large co-regulatory complexes, and the role of the polymerase machinery in setting interactions between regulatory elements.
Improve disease outcomes of developmental disorders
Our aims are designed to identify genomic regions as well as nuclear factors that may be hypersensitive to sequence variation. Not only will this enable early diagnosis, it may also help design targeted therapies. The development of animal models can then be used to directly test such therapies.
Approach
We generate allelic series of genetically engineered mouse models with different types of perturbations of chromatin structure, and employ advanced genomics techniques to molecularly phenotype these mutants. We complement in vivo work with cell culture-based differentiation, where we employ our modifications of massively parallel reporter assays to probe the functional role of different regulatory elements in controlling gene expression under distinct developmental and genomic contexts.
Examples of our work
CTCF loops promote and repress enhancer contacts
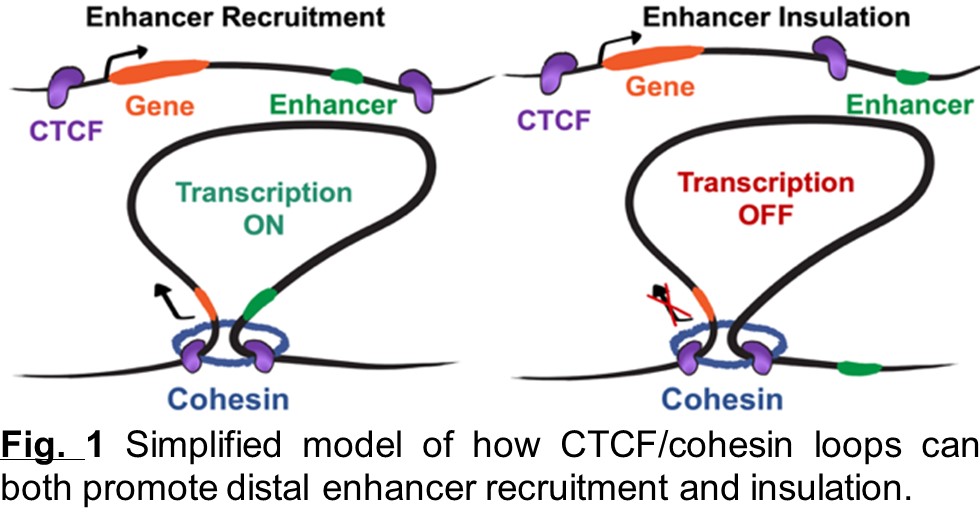 These loops form through the ability of the cohesin complex to extrude DNA and of CTCF to retain extruding cohesin. CTCF loops can facilitate contacts between enhancers and genes but also ensure that enhancers cannot activate genes outside of their loop. However, it isn’t yet clear how these models are generalizable to all genes and how the impact of CTCF-cohesin loops on gene regulation may be affected by different types of genes, enhancers, and loops.
These loops form through the ability of the cohesin complex to extrude DNA and of CTCF to retain extruding cohesin. CTCF loops can facilitate contacts between enhancers and genes but also ensure that enhancers cannot activate genes outside of their loop. However, it isn’t yet clear how these models are generalizable to all genes and how the impact of CTCF-cohesin loops on gene regulation may be affected by different types of genes, enhancers, and loops.
Transcriptional insulation by CTCF loops is modulated by enhancer type and boundary strength
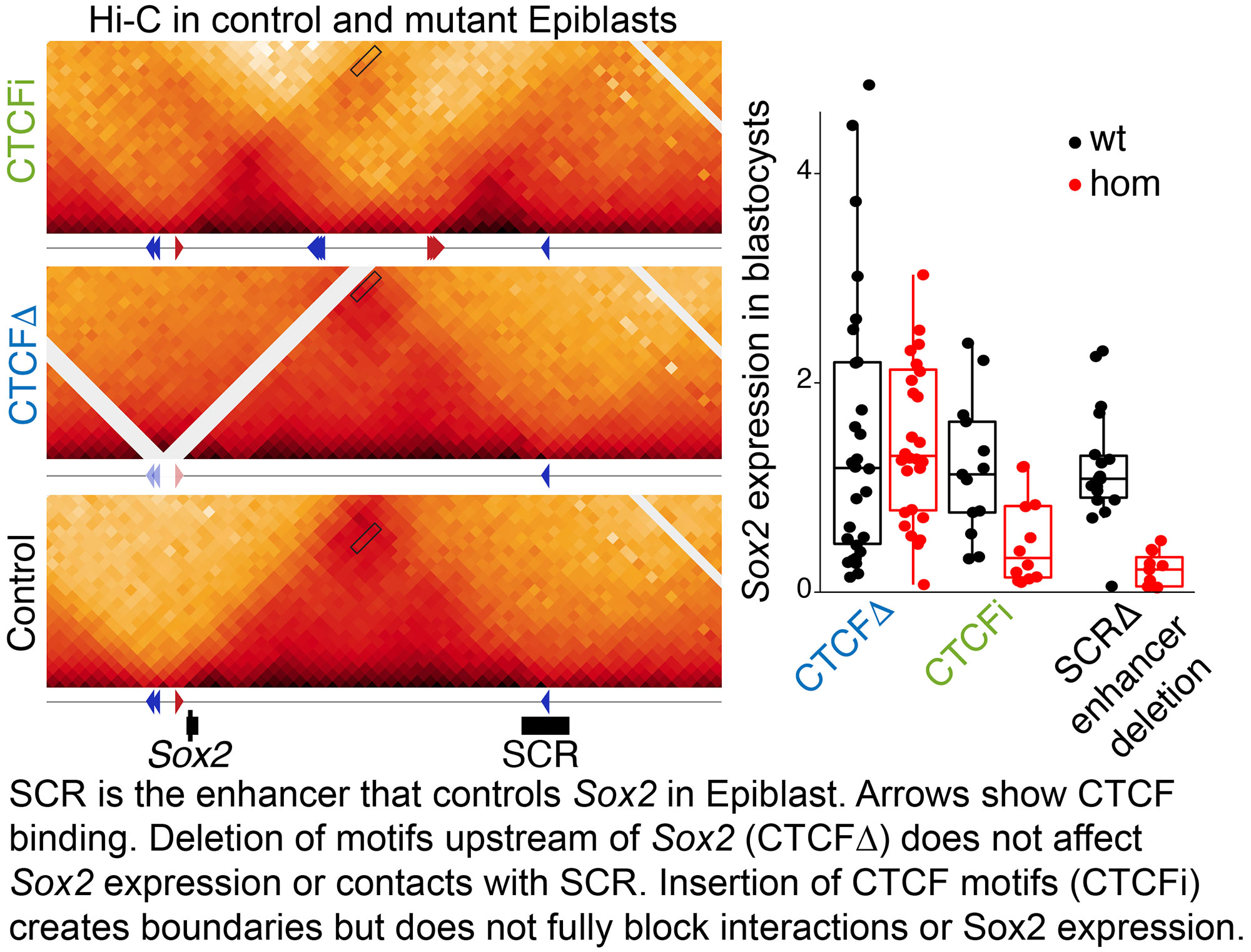 SOX2 is a Transcription Factor important for Epiblast specification, neurogenesis and tracheoesophageal separation. Its locus is bound by CTCF directly upstream of its promoter, which suggest that CTCF-cohesin loops could be used to recruit its different tissue specific enhancers. Surprisingly, we showed (Chakraborty Nature Genetics 2023
SOX2 is a Transcription Factor important for Epiblast specification, neurogenesis and tracheoesophageal separation. Its locus is bound by CTCF directly upstream of its promoter, which suggest that CTCF-cohesin loops could be used to recruit its different tissue specific enhancers. Surprisingly, we showed (Chakraborty Nature Genetics 2023 ) that Sox2 recruits its enhancers through CTCF independent mechanisms. We also generated an extensive series of mouse mutants with insertions of ectopic loops between Sox2 and its enhancers. By comparing different types of insertions, we determined that the highest transcriptional insulation is achieved when extruding cohesin complexes are blocked by CTCF from both directions of an active enhancer-promoter pair.
In contrast to the Epiblast and Neural Tissues, Sox2 was undetectable in the anterior foregut of mutants carrying the strongest boundaries, and these animals fully phenocopied loss of SOX2 in this tissue. The blastocyst and neural Sox2 enhancers are found within clusters of high density of TF motifs and recruit high levels of coregulatory complexes. In contrast, in the anterior foregut, Sox2 enhancers are spread through large genomic stretches at lower density. We therefore proposed that these different enhancer configurations determine the ability of enhancer–promoter interactions to bypass CTCF loops. 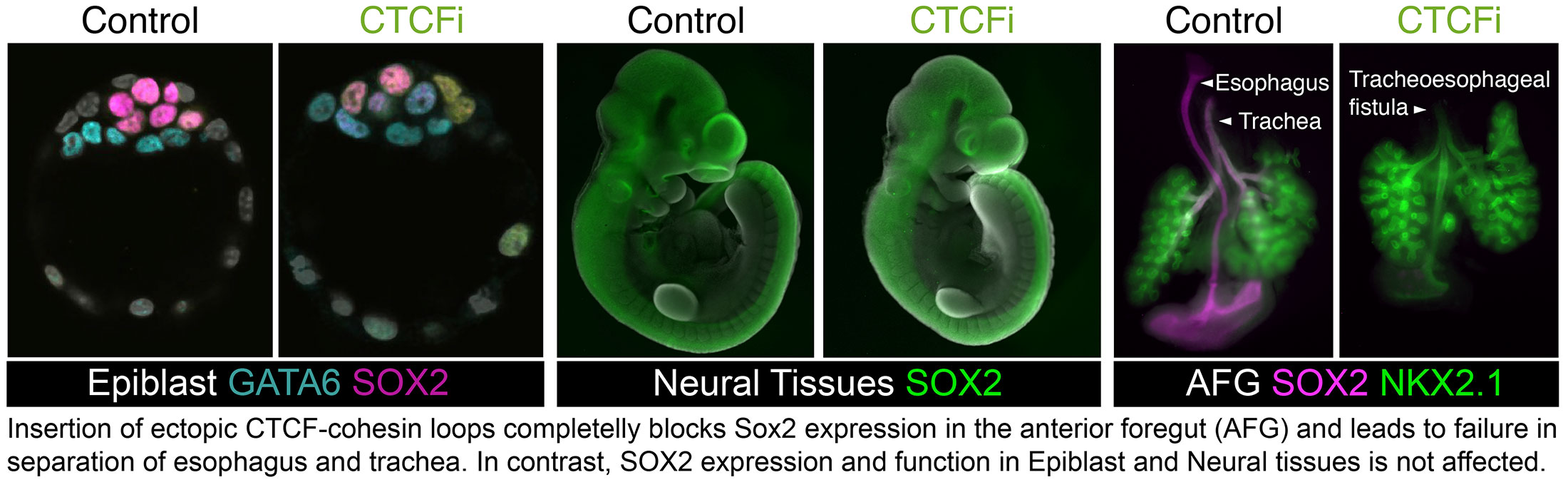
Loss of a single CTCF motif can severely dysregulate gene expression and mouse development
We hypothesized that in loops with multiple developmental regulators, CTCF could be particularly important to ensure tight tissue-specific expression. To test this, we studied a 63kb loop harboring three essential FGF ligand genes—Fgf3, Fgf4, and Fgf15. Removing the most centromeric boundary (C1-C4) of the domain led to striking lethal phenotypes. In fact, this is the first demonstration that heterozygous loss of a CTCF boundary can cause fully penetrant perinatal lethality. These embryos developed encephalocele, together with ectopic FGF expression in the brain. Surprisingly, we showed that loss of the single CTCF motif oriented towards Ano1 (C1) fully recapitulates these phenotypes but deleting the three motifs pointing to the FGF genes (C2-C4) did not. This study (Chakraborty Developmental Cell 2025 ) revealed that sequence variants—as small as a single CTCF motif—at particular domain boundaries can have a surprisingly outsized effect and must be considered as potential sources of gene dysregulation in development and disease. This is the first mouse model of encephalocele—a birth defect that affects 1:10000 human births. We are currently using it to investigate the etiology of this disease and potential therapies.
Rewiring of Regulatory Interactions
 As cells progress through their differentiation trajectories to become more and more specified, regulatory interactions between enhancers and promoters have to be remodeled and re-established. We are very interested in understanding the dynamics of this reorganization, the mechanisms that drive it, and how its importance to specification of cell fates may change throughout development. For example, during specification of the Primitive Endoderm we have shown (Thompson Nature Communications 2022
As cells progress through their differentiation trajectories to become more and more specified, regulatory interactions between enhancers and promoters have to be remodeled and re-established. We are very interested in understanding the dynamics of this reorganization, the mechanisms that drive it, and how its importance to specification of cell fates may change throughout development. For example, during specification of the Primitive Endoderm we have shown (Thompson Nature Communications 2022 ) that the co-bound state of NANOG and GATA6 in ICM cells, is followed by NANOG eviction and repression of Epiblast genes. This is accompanied by quick remodeling of chromatin and enhancer-promoter contacts. This dynamic rewiring of the genome is not unique to the Blastocyst. We have recently shown that a specialized cell type of our immune system, Dendritic Cells, also rearrange their genome throughout differentiation (Kurotaki PNAS 2022
).
 BACK TO TOP
BACK TO TOP